1. Background
Ichthyosis refers to a group of skin disorders that can occur in both hereditary and acquired forms in individuals. This disorder results in a deficiency in the differentiation of keratinocytes and an abnormal structure of epidermal cells (1). Ichthyosis is classified into three groups based on genetic factors:
- Autosomal Dominant Group: This group includes Ichthyosis vulgaris, Lamellar ichthyosis, Refsum syndrome, and Sjögren-Larsson syndrome. These conditions follow an autosomal dominant inheritance pattern.
- Gender-Dependent Dominant Group: A type of ichthyosis that follows a gender-dependent dominant pattern.
- Autosomal Recessive Group: This group includes Ichthyosis Vulgaris bullosa and, rarely, Hystrix ichthyosis. These conditions follow an autosomal recessive inheritance pattern (2).
Despite the existence of various disease types, common symptoms in all forms of ichthyosis include itching, recurrent infections, sweating disorders (hypohidrosis) accompanied by heat intolerance, and various ocular, auditory, and nutritional complications (3, 4). In recent years, significant advances have been made in understanding the pathophysiology of ichthyosis (5, 6). Studies indicate that mutations in more than 40 genes can lead to the ichthyosis phenotype (7, 8). However, diagnosing the disease based solely on clinical symptoms and histopathological lesions is not precise enough to identify the molecular cause (9).
Next-generation sequencing (NGS) technology, specifically whole-exome sequencing (WES), makes it possible to sequence all coding protein regions as well as exonic-intronic boundary regions. This high-efficiency technique enables the identification of the molecular basis of many genetic disorders (10, 11).
Our research aims to explore the molecular dimensions of ichthyosis using cutting-edge WES technology. By scrutinizing carriers genetically, we can identify the underlying factors of genetic skin disorders and inhibit the propagation of harmful mutations to succeeding generations. Crafting an exome test panel covering all pertinent genes linked to these skin ailments enables a precise, swift, and economical molecular diagnosis.
2. Objectives
This represents a notable advancement in our pursuit to analyze and comprehend the genetic complexities of skin disorders efficiently and with the utmost precision possible.
3. Methods
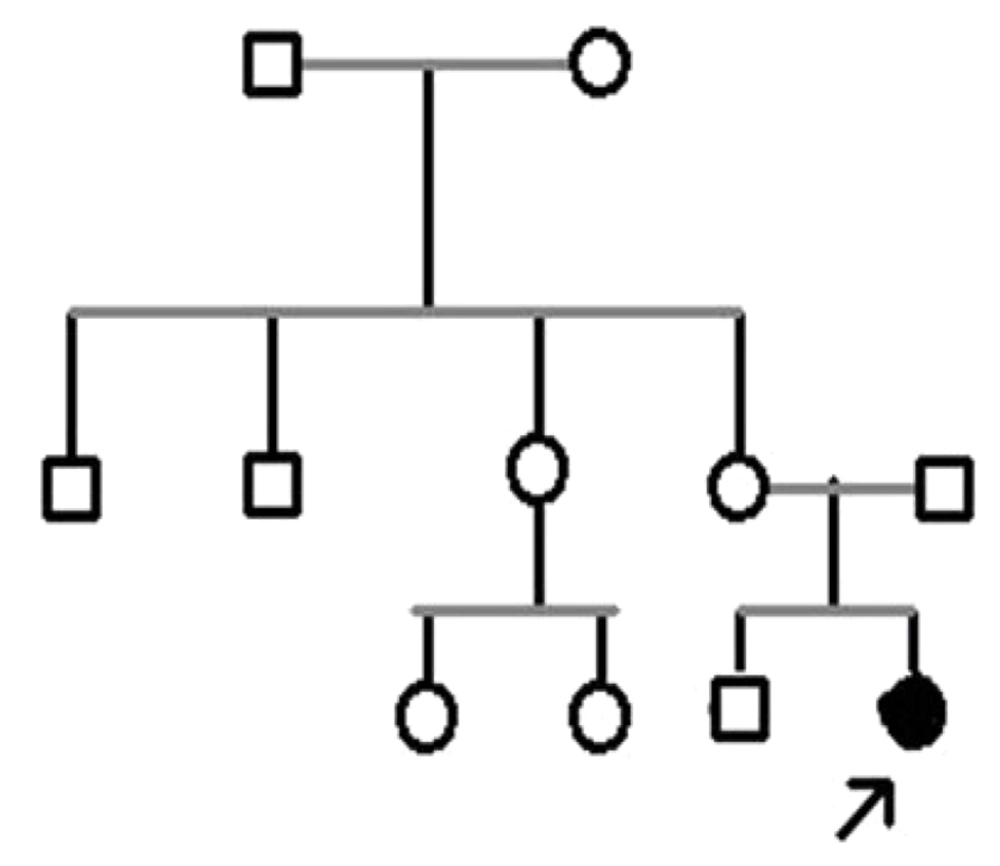
In this cross-sectional study, we investigated a family with a diseased child and seven of their relatives who visited the Noorgene Medical Genetics Laboratory during the years 2022 - 2023. Entry into the study required meeting specific criteria, including confirmation of clinical manifestations related to ichthyosis by a specialist physician and obtaining consent from the research participants. After collecting clinical data and providing genetic counseling, we constructed a family pedigree (Figure 1).
We collected 5 milliliters of the patient's peripheral blood in a 0.5% EDTA tube and performed DNA extraction using the salting-out method. Quantitative and qualitative assessments of DNA quality were conducted using a nanodrop device and electrophoresis on agarose gel. Genome sequencing was carried out using the Illumina HiSeq 2500 platform. The analyzed data were then validated against the pedigree pattern using the Sanger sequencing standard method.
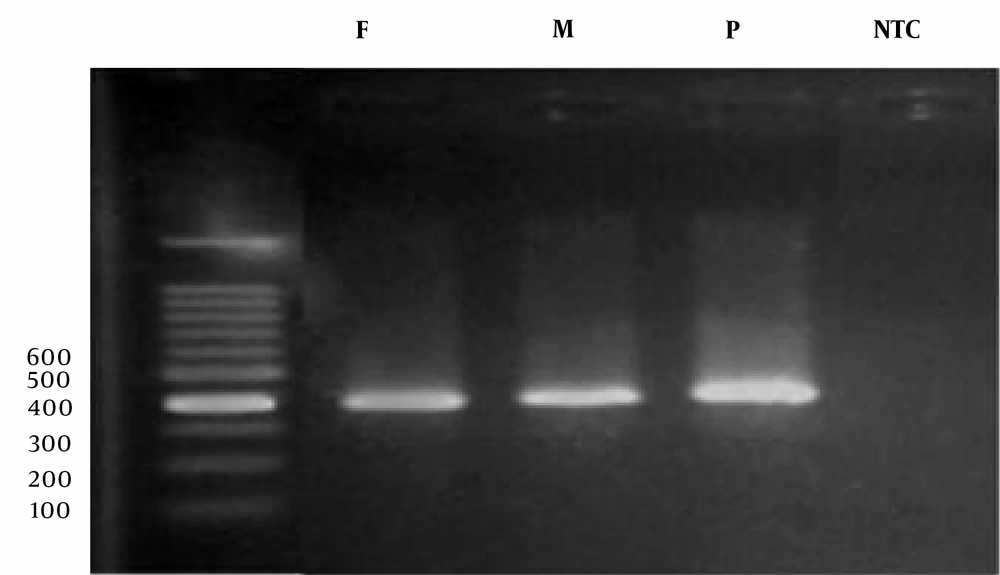
For PCR execution, we added 10 microliters of Red Mix solution, 8 microliters of double-distilled water, 2 microliters of the DNA sample, and 0.5 microliters each of forward and reverse primers to the reaction tube. The primers used in the research are detailed in Table 1. The PCR products underwent electrophoresis (Figure 2). Finally, the PCR products were sequenced using the ABI 3130XL sequencing instrument, and the results were analyzed using Chromas software on the NCBI and ENSEMBL websites.
| Primer | Sequence |
|---|---|
| NIPAL4-EX6-F-399 | GGG CAA AGG AAT ATC CTC ATC TAC |
| NIPAL4-EX6-R-399 | GCA GCT GAT GTC CAG GTC TT |
4. Results
In this study, we examined a family from Khuzestan province along with seven of their relatives. After analyzing the results and constructing the pedigree, we assessed the type of mutation present in the family. Within this family, a diseased daughter exhibiting phenotypic symptoms of ichthyosis was identified and confirmed by a dermatologist. The pedigree analysis revealed that other family members and relatives were asymptomatic.
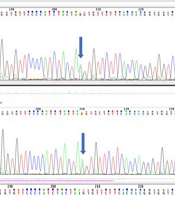
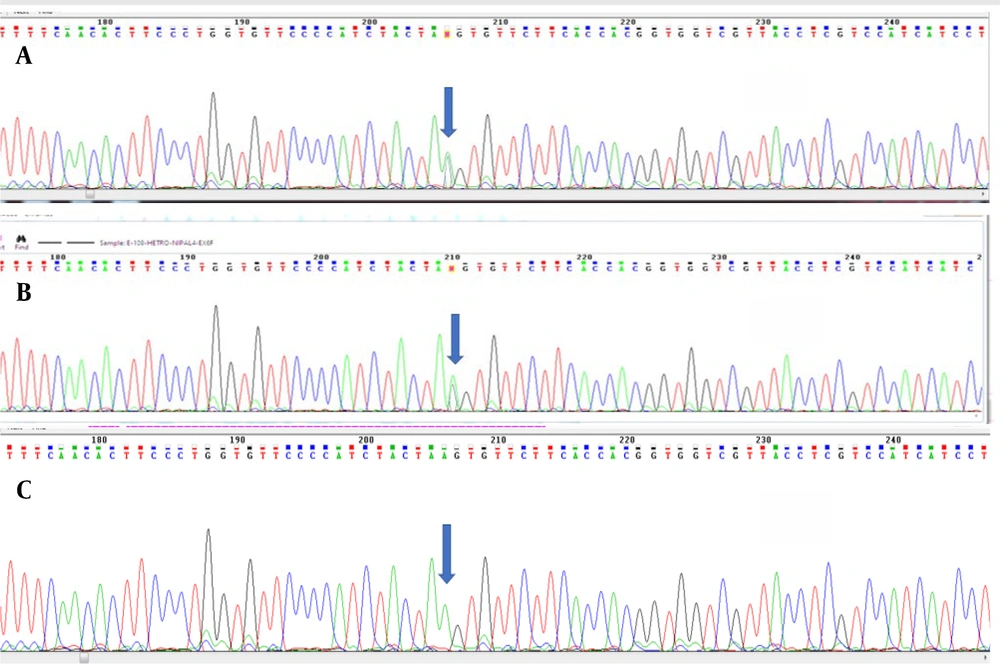
Whole Exome Sequencing (WES) results identified 256,987 variants, which were further filtered based on zygosity and allele frequency in the population. This process led to the identification of a variant as a candidate molecular factor in the disease. The specific mutation, NM_001099287.2: Exon 6:c.C897A:p.Ter361Ter, is located in the NIPAL4 gene at the chromosomal location 5q33. This variant, situated in exon 6 at codon 1083, is normal in the CC state, diseased in the AA state, and heterozygous in the AC state, indicating carrier status for the individual (see Figure 3).
5. Discussion
Ichthyoses represent a heterogeneous group of keratinization disorders characterized by genotypic and phenotypic abnormalities, resulting in continuous shedding of the skin due to improper epidermal layering (12). Given that ichthyoses follow several inheritance patterns, the candidate gene identified for the studied family is NIPAL4, inherited in an autosomal recessive manner. Autosomal Recessive Congenital Ichthyosis (ARCI) is caused by functional defects in epidermal barrier components, including the intercellular lipid barrier, cornified cell envelope, and keratin breakdown products, leading to chronic scaling and skin roughness. In most cases of ARCI, skin dryness, fissures, and occasional itching are also reported. ARCI can be particularly life-threatening in the first year of life due to severe skin inflammation, hypothermia, and dehydration (13). This disease, which can be inherited as a distinct trait or as part of a syndrome in combination with other abnormal manifestations, has a prevalence of less than 1 in 200,000 and manifests due to mutations in several genes (14). Based on our study, comprehensive genetic exploration focusing on the NIPAL4 family of genes involved in skin diseases has not been conducted in Iran to date, with only limited studies addressing the investigation of different inheritance patterns of the disease. Next-Generation Sequencing (NGS) techniques are highly valuable tools for detecting genodermatoses (disorders that can manifest specifically in the skin), including ichthyoses (15). Between 2009 and 2019, NGS has aided in identifying 166 gene relationships related to skin diseases and 63 cases of complex genodermatoses caused by multiple gene mutations (16). In 2018, Sitek et al. proposed the use of Whole Exome Sequencing (WES) as the primary diagnostic method for ichthyosis after studying 34 suspected cases over a three-year period (17). Therefore, it appears that by employing new techniques, a fundamental revolution in understanding and diagnosing skin disorders, including ichthyoses, is underway.
Charfeddine et al.'s in silico analysis demonstrated that mutations in the 3D domain structure lead to NIPAL4 protein instability and impaired Mg2+ transport, pointing to the potential role of the NIPAL4 gene in the development and maintenance of epidermal barrier function. Collectively, the results of their study strengthened NIPAL4 as the main candidate gene for the EKV-like Autosomal Recessive Congenital Ichthyosis (ARCI) phenotype (18).
The use of acellular scaffolds can be an important step in the treatment of skin diseases such as ichthyosis (19).
This research aims to effectively identify new mutations in the NIPAL4 gene, responsible for a familial skin disease, through the use of advanced and effective techniques. Identifying disease-causing mutations will help reveal the genetic structure of this disease and subsequently enhance preventive and counseling methods significantly. Furthermore, studying the genes associated with genetic syndromes will help doctors better understand the systems involved in the disease (20).
By conducting further investigations on a larger number of families, it will be possible to determine the prevalence and types of gene mutations involved in skin diseases in Iran. This could lead to the design of a specialized panel for ichthyoses, which would greatly assist in diagnosing the disease, preventing new cases within families, and improving overall community health.
5.1. Conclusions
Our study identified the NIPAL4 gene as a candidate gene inherited in an autosomal recessive manner, highlighting its potential involvement in Autosomal Recessive Congenital Ichthyosis (ARCI). While comprehensive genetic exploration focusing on the NIPAL4 gene family in Iran remains limited, advancements in Next-Generation Sequencing (NGS) techniques offer promising avenues for identifying and understanding genodermatoses like ichthyoses. The use of Whole Exome Sequencing (WES) has shown promise as a primary diagnostic tool, revolutionizing our ability to elucidate the genetic basis of skin diseases.