1. Background
The emergence of antibiotic resistance is a global public health problem. Gram-negative bacterial resistance is of particular importance, since there is a dearth of novel antibiotics directed against these organisms. The clinical utility of carbapenems and the agents of the last resort against multi-drug resistant Enterobacteriaceae are threatened with the growing incidence of pan resistant isolates (1, 2). Different species of Klebsiella genus are responsible for a wide variety of diseases in humans. Klebsiella pneumoniae has been associated with various ailments such as urinary tract infections, septicemia, diarrhea, and other diseases (3). In addition to being the primary cause of respiratory tract infections, K. pneumonia is commonly involved in acute pyelonephritis in pregnant women with urinary tract abnormalities such as urolithiasis, hydronephrosis or congenital deformities. Several reports, especially from the Asia Pacific region and the United States, have also shown that this pathogen has become the predominant cause of liver abscess (4, 5). Tetracycline and other antibiotics have been frequently used to treat diseases, but unfortunately repeated use of these compounds has resulted in the development of resistant strains. Tetracycline resistance in bacteria is mediated by four mechanisms: efflux, ribosomal protection, enzymatic inactivation, and target modification (6). At present, 23 genes encoding efflux pumps and 11 genes encoding ribosomal protection proteins, not including the recently described mosaic tetracycline resistance genes have been reported (7). In prior clinical surveys, tetB gene was identified as the most prevalent tetracycline resistance determinant with a wide host range due to the fact that it resides on highly mobile genetic elements that readily transfer between different bacterial genera (8). The gene tetA is located on conjugative plasmids of different incompatibility groups.
2. Objectives
The current study aimed to detect tetA and tetB genes in Klebsiella Pneumoniae isolated from clinical samples.
3. Materials and Methods
In the present cross-sectional study, a total of 30 K. pneumoniae strains were isolated from urine cultures of hospitalized patients in Mir Hospital (Zabol, south east of Iran) who had urinary tract infections from 2011to 2012. All samples were collected aseptically from the patients and plated right after the collection. Identification of all causative microorganisms was performed by standard microbiologic methods (9).
3.1. Agar Disk Diffusion Assay
The susceptibility of all applied antibiotics was carried out using disc diffusion method on Mueller-Hinton agar as recommended by Clinical and Laboratory Standards Institute (CLSI) (10). Briefly, K. pneumoniae isolates were grown overnight on blood agar, and colony suspension was prepared equal to the turbidity of a 0.5 McFarland standard. The suspension (100 μL) was spread over a Mueller-Hinton agar plate and discs of the selected antibiotics were placed aseptically on the surface of inoculated media. Antibiotics concentrations were as follow: cefixime (30 μg), tetracycline (30 μg), erythromycin (15 μg) and ceftazidime (30 μg).
3.2. Amplification of Genes
The colonies of K. pneumoniae were suspended in TE (Tris + EDTA) buffer and their DNA was extracted by simple boiling method (11). The polymerase chain reaction (PCR) method was performed to detect tetA and tetB genes as described previously with minor modifications, using specific pair of primers (Table 1).
| Primer | Sequence | Annealing Temperature |
|---|---|---|
| tetA | - | - |
| F | GTGAAACCCAACATACCCC | 58 |
| R | GAAGGCAAGCAGGATGTAG | 60 |
| tetB | - | - |
| F | CCTTATCATGCCAGTCTTGC | 60 |
| R | ACTGCCGTTTTTTCGCC | 52 |
aAbbreviations: F, forward; R, reverse
The PCR mixture consisted of 10 pmol of each primers, 10 ng DNA sample, 1.5 mM MgCl2, 0.2 mM each dNTP, and 5 U Taq DNA polymerase (Cinagen, Iran) in a total volume of 50 μL of PCR reaction.
4. Results
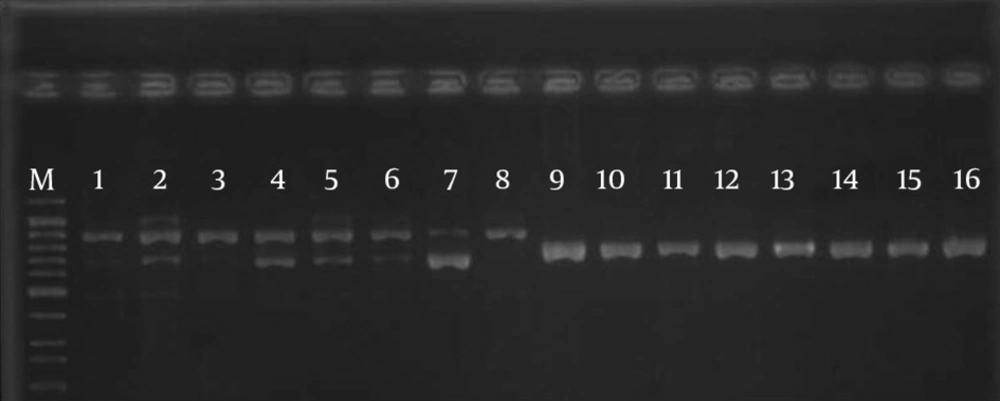
Overall, K. pneumoniae isolates were resistant to the agents including erythromycin (70%), cefixime (53.3%), tetracyclin (50%) and ceftazidime (36.6%) (Table 2). The amplification of tetA and tetB of K. pneumoniae revealed that all of the isolates harbored these genes (Figure 1).
| Ceftazidime | Cefixime | Erythromycin | Tetracycline | |
|---|---|---|---|---|
| Sensitive | 16 (53.3) | 10 (33.3) | 2 (6.6) | 13 (43.3) |
| Intermediate | 1 (3.3) | 2 (6.6) | 5 (16.6) | 1 (3.3) |
| Resistant | 11 (36.6) | 16 (53.3) | 21 (70) | 15 (50) |
aData are presented as No. (%)
5. Discussion
The K. pneumoniae isolates in the study were resistant to erythromycin (70%), cefixime (53.3%), tetracyclin (50%) and ceftazidime (36.6%). In the study conducted by Derakhsan et al. more than half (17/31) of K. pneumoniae isolates showed high resistance to different antibiotics including amoxicillin-clavulanic acid, cefotaxime, ceftriaxone, aztreonam, and ceftazidime (12). All of the isolates in the current study harbored tetA and tetB genes. In the study by Tuckman et al. a total of 452 tetracycline-resistant and nonduplicate isolates were positive by PCR for at least one of the six examined tetracycline resistant determinants (13). In the latter study, 32% and 26% of isolates were positive for tetB and tetA respectively, whereas tetC, tetD, tetE, and tetM, were collectively found in 4% of isolates (13). The tetracycline resistance is a common and developing aspect of resistance among the other bacteria. In the study by Skockova et al. for example, 102 Escherichia coli isolates were examined and about half (49.0%) of these isolates showed resistance to tetracycline. Antibiotic resistance and the corresponding gene(s) show time and geographical dependency. The most common gene detected in tetracycline-resistant isolates from 2010 to 2011 was tetA (81.3%), while tetB was most often (86.5%) found in isolates from 2005 to 2006 (14). The tetracycline resistance genes (including tetA and tetB) have been detected from clinical and non-clinical samples. Koo and Woo have identified tetA (52.4%) followed by tetB (41.3%) as the most frequent genes in tetracycline-resistant E. coli isolates from meat and meat products (15). In the study by Menggen et al. antimicrobial resistance genes, carried by 30 Salmonella isolates, were detected and common genes included tetC (60%, 18/30), cat1 (43.3%, 13/30) and tetA (40%, 12/30) (16). These studies provide a picture of the tetracycline resistance genes burden among clinical and nonclinical K. pneumoniae isolates, as well as the utility of the novel broad-spectrum agent, and tigecycline against these pathogens. Moreover, these results support the general approach of reengineering the existing antimicrobial agents with acceptable safety profiles to evade the resistance mechanisms posed by bacterial pathogens.
In conclusion, resistance to tetracycline and other antibiotics and the presence of various resistance determinants in K. pneumonia strains is an alarming sign in Zabol area. Wide and inappropriate use of antibiotics may play an important role in resistance development. The current study strongly recommends limiting the consumption of antibiotics including tetracycline. Further studies should be conducted in order to find out the extent of the problem in other areas.