1. Background
Blood coagulation disorders are results of deficiency in coagulation factors activity. Coagulation factor XIII (FXIII) has an essential role at the end of coagulation cascade and the stabilization of fibrin clot. FXIII is a zymogen and its active form is a transglutaminase which catalyzes an acyl transfer reaction and cross-links between two polypeptide chains forms through an γ-glutamyl lysine link (1). FXIII is heterotetramer (A2B2) in plasma consisting of two catalytic A subunits and two B subunits; the B subunit act as a carrier that protects and fixates the A subunits (1). The FXIII-A subunit is encoded by a gene on chromosome 6p24-25 as a protein consisted of 731 amino acid (2). It comprises five domain including the activation peptide (residues of 1-37), β-sandwich (residues of 38-183), central domain or catalytic core region (residues 184-515), and β-barrel 1 and 2 (residues 516-627 and residues 628-731, respectively) (3). The FXIII-B subunit is encoded by a gene located on chromosome 1q31–32.1. This part spans 28 kb and comprises 12 exons and 11 introns (4). FXIII deficiency is a rare bleeding disorder; its frequency is one to three per million populations and is transmitted in an autosomal recessive manner. It is usually more prevalent in areas with high rate of consanguineous marriages (1, 5). FXIII deficiency is associated with umbilical cord bleeding, prolonged wound healing, recurrent spontaneous miscarriage, and spontaneous intracranial hemorrhage (6). After FVIII deficiency, FXIII is the most common bleeding disorder in Sistan and Balouchestan Province in the southeast of Iran (7). up to this date, 133 different mutations were reported in FXIII gene in which 114 occurred in FXIII-A subunit and only 19 mutations occurred in FXIII-B subunit. Missense mutations are the most prevalent mutations in FXIII gene (1, 6, 8, 9). Previous studies reported few mutations in patients with FXIII hemophilia in Iranian population and Arg77His and Trp187Arg were the most common mutations (10).
2. Objectives
The aim of this study was to determine the c559T > C FXIIIA Genotype frequency in patients with FXIII hemophilia by T-ARMS-PCR.
3. Patients and Methods
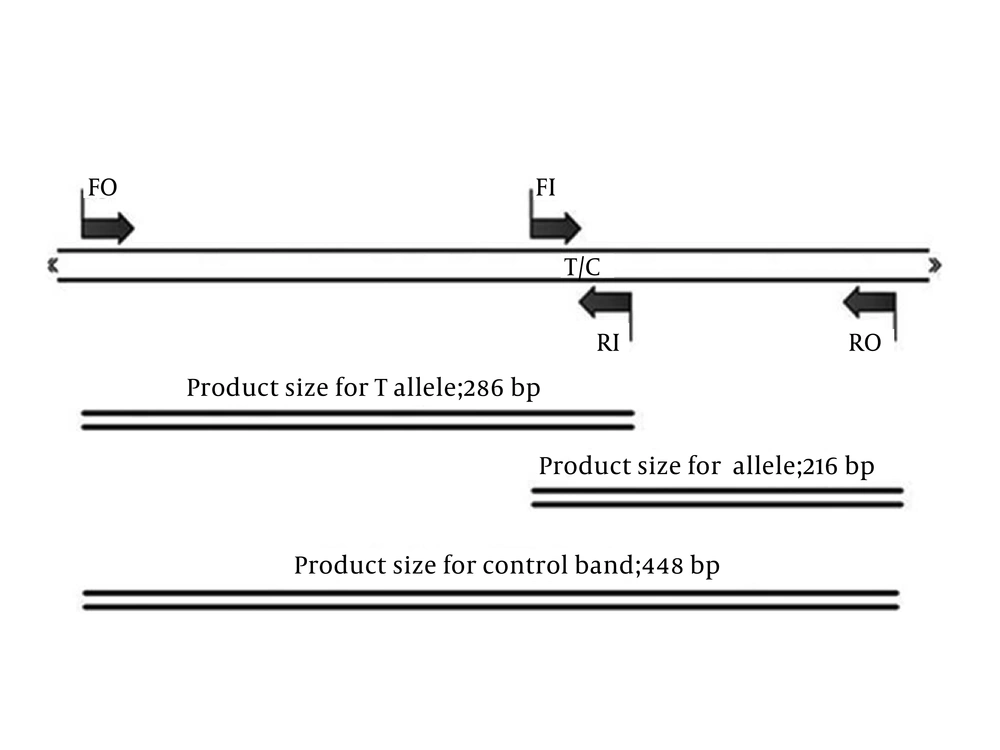
This study was conducted on 180 patients with FXIII hemophilia who were referred to Ali Asghar Hospital with clinical symptoms of coagulation disorder and had normal results for prothrombin time (PT), Partial thromboplastin time (PTT), bleeding time (BT), fibrinogen level, and platelet count tests. The most common screening tests for FXIII are based on solubility of fibrin clot in solutions (2% acetic acid or 1% monochloracetic acid or 5 Mol urea). In abnormal cases, FXIII level was measured by quantitative factor assays and genotyping was performed. This study was approved by the Ethics Committee of Tehran University of Medical Sciences and an informed consent was obtained from each participant. Genomic DNA was extracted from peripheral blood (11) and c559T > C mutation was detected by considering that this mutation was the most common mutation reported by T-ARMS-PCR (9, 12-15). We designed four primers to determine the c559T > C mutation of FXIIIA. We used, two external primers (forward outer (FO): 5′-TCTTCTCTCTTCTCATAGGTCGCTACC-3′, reverse outer (RO): 5′-CACCACCACACCCATCTAATTTTTAA-3′) and two internal allele-specific primers (forward inner (FI) [C allele]: 5′-AACAGACACGTACATTCTCTTCAATCATC-3′, reverse inner (RI) [T allele]: 5′-AATGTCTGCCTCTTACCTTCACACAA-3′). The products size were 216 bp for C allele, 286 bp for T allele, and 448 bp for the two outer primers (control band), as is schematically shown in Figure 1.
Each PCR contained 10 mmol Tris-HCl, pH = 8.3, 1.5 mmol MgCl2, 200 μMol each dNTP, 10 pmol each primer, 200 ng genomic DNA, and 0.5 U Taq DNA polymerase in a final volume of 25 μL. The PCR conditions included 31 cycles, each lasting 60 s, with the following temperatures: denaturation at 95˚C, annealing at 60˚C, and extension at 72˚C with a final step at 72˚C for five minutes. Reaction products were separated on a 3% agarose gel containing ethidium bromide.
4. Results
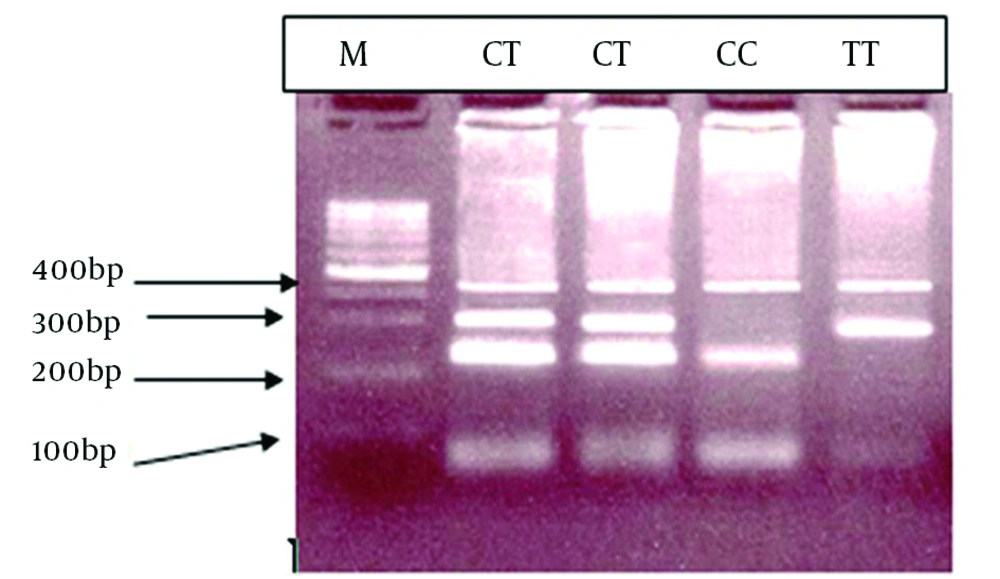
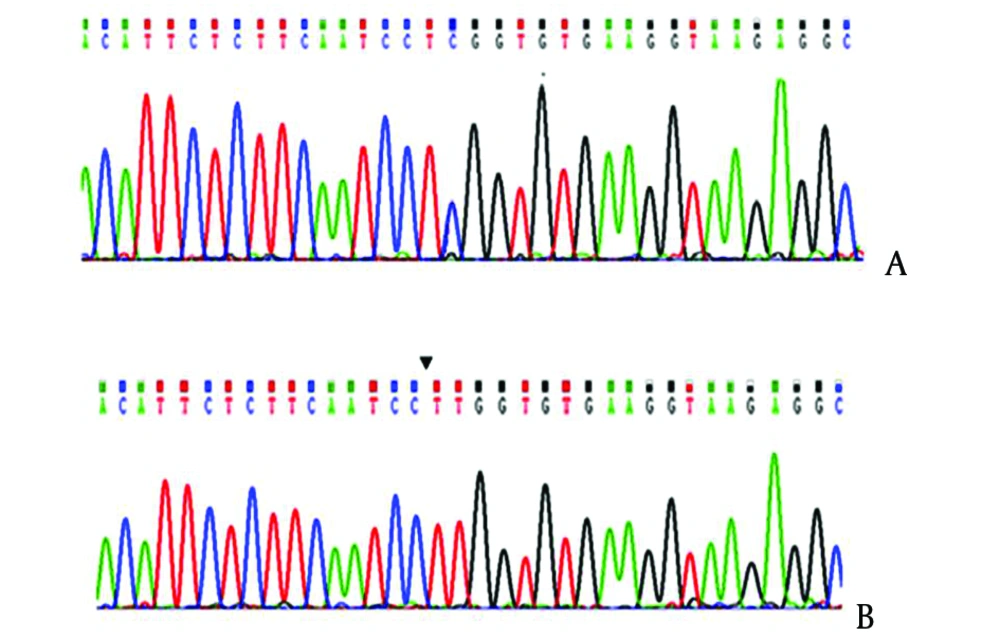
We evaluated 180 patients with hemophilia, 174 (96.6%) patients had substitution of C to T and change of Tryptophan to Arginine in the form of homozygote. The T-ARMS-PCR method was effectively applied for genotyping C559T > C mutation of the FXIIIA. The genotypes determined by this method is shown in Figure 2, and conformities sequencing are illustrated in Figure 3.
5. Discussion
FXIII deficiency is a rare autosomal recessive disorder with a higher prevalence rate in Sistan and Balouchestan province than the worldwide rate. This high prevalence might be due to the consanguineous marriage as more 70% of marriages are consanguineous in this province (10). In this area, the number of patients diagnosed were Significantly increased from 45 in 2006 to 205 cases in 2012, (16). Sistan and Balouchestan province with 2,700,000 population had about 200 FXIII deficient known cases (1 per 13,500 population), which is 70 times higher than the global average. The number of FXIII deficient cases were few in the other parts of the world, for example, in UK it was 26 cases (7) and 72 cases were recognized in a vast study carried out in European Coagulation Disorders Research Center (1996) (17). Frequency ratio of factor XIII deficiency to blood coagulation disorders was reported in 62% Caucasians, 6% in African-America, 19% in Latinos, 3% in Asian (7) whereas Factor XIII deficiency is the second most common blood coagulation disorders in southeast of Iran (10). In the present study, we showed that c.559T > C mutation was the most common mutation (more than 95%) in FXIII gene in Sistan & Balouchestan province. T-ARMS-PCR is a quick and easy technique for determination of the mutation (18). This method was easily applied for genotyping the Trp187Arg in FXIIIA gene and the determined genotypes were confirmed by the sequencing genotypes. The parents’ DNA showed heterozygous pattern. One of the advantages of this method is that efficiently amplifies both wild type and the mutated alleles with a control fragment in a single PCR tube. A large number of mutations have been reported according to different databases for FXIII gene (http://www.hgmd.cf.ac.uk and http://www.f13-database.de) in different parts of the world. Anwar et al. reported four mutations codon 295; G → A at position -246 upstream of exon 1; T → C and C → T at positions -23 and -24, respectively, in intron nine in patients with FXIII hemophilia who originated from the North of Pakistan, the neighboring country of this province (19). The common mutations in five different subunits of FXIII-A gene are shown in Table 1. Previous studies reported Arg77His and Trp187Arg as the most common mutations (10).
| Type of Mutations and Region | Beta Sandwich | Core | Beta Barrel 1 | Beta Barrel 2 | Activated Peptide |
|---|---|---|---|---|---|
| Missense | Asn60Lys, Arg77Cys | Pro186Leu,Trp187Arg,Gly210Arg, Gly215Arg, Leu235Arg, Met242Arg, Arg252Ile, Arg260Cys, Arg260His, Arg260Leu, Gly262Glu,Ser263Phe, Trp283Cys, Pro289Arg, Ser295Arg, Val316Phe,Ala318Val,Arg326Gln,Leu354Pro, Ala378Pro, Arg382Ser, Ala394Val,Thr398Asn,Arg408Gln, Ser 413Trp, Val414Phe,Gly420Ser, Leu498Pro,Gly501Arg | Arg540Gln | Gly592Ser, Arg611His | |
| Arg77His, Arg78Cys | Asn541Lys | Leu660Pro, Leu667Pro | |||
| Met159Arg, Tyr167Cys | Gly562Arg | Asp668Gly, Arg703Trp | |||
| Ser708Asn, His716Arg | |||||
| Non sense | c.210T>G, c.514C>T | c.979C>T, c.1201C>T, c.1171G>T, c.1326C>A | - | c.1984C>T, c.1994G>A | |
| c.523 C>T | c.2075G>A | ||||
| Insertions/deletions | (−7 to −20) insTT | c.599–600delAA, c.689delA, Exon 5 del, ins T866, c.709delG | c.1652del10bp | c.2002–2003delCT | 33bpdel |
| c.291–296insTCGTCC | c.869insC, c.1030–1032delAAT, c.1201insC, 127067del33 | c.2066del C, Exon 15 del | c.27delT | ||
| c.249–261del13bp | c.1392–1395delAATT, c.1405–1408delCAAA, Gross Deletion(>100 kb) | ||||
| c.397insG | |||||
| c.499–512del14bp | |||||
| Splice site | c.131–132delAG | IVS5−1 G>A, IVS7+1 G > A, c.1111T>G, IVS10+1G, c.2045G>A | |||
| c.319GNT, IVS3+6 T > C, IVS3+5 G>A |
Considering the high rate of consanguineous marriages, the great homogeneity in the population and mutations was seen in this area. The sequencing change in this mutation is TGG → CGG and is related to a tryptophan to arginine substitution at residue 187 (20). Trp187 in FXIIIA is conserved in all known transglutaminases (21). In the FXIIIA subunit, residue of Trp187 is located in the direction to the surface of the protein between the catalytic core and the b-sandwich domain (22) and substitution of this residue with arginine would increase steric clashes between the Arginine side chain atoms and the contiguous atoms from the neighboring residues (20). In sum, C559T > C mutation was a risk factor for predisposition to FXIIIA deficiency in this part of Iran. It seemed that this province had the highest prevalence rate of FXIII deficiency in the world. With respect to the high incidence of FXIIIA gene deficiency, an appropriate screening system to identify the carriers is essential. Due to limited mutations in this area, T-ARMS-PCR with a sound speed, accuracy, sensitivity, as well as cheapness can be used alongside the common screening tests.