Dear Editor,
Mitochondrial dysfunction is a hallmark of metabolic syndrome (MeS), represented by a compendium of metabolic abnormalities that include insulin resistance, obesity, hyperlipidemia, hypertriglyceridemia and hypertension, which result in cardiovascular (CVD) and non-alcoholic fatty liver disease (NAFLD). Whether mitochondrial dysfunction is a cause or consequence of these metabolic abnormal conditions is unknown. However, since mitochondria have a central role not only in energy production through the metabolism of fatty acids and carbohydrates, but is also key in the synthesis of heme, urea, steroids and pyrimidine, its alteration can produce metabolites that can exacerbate these metabolic abnormalities and greatly increase the progression of CVD and NADFLD. Thus, understanding mitochondrial function and regulation will provide insight into the maladies associated with MeS (1).
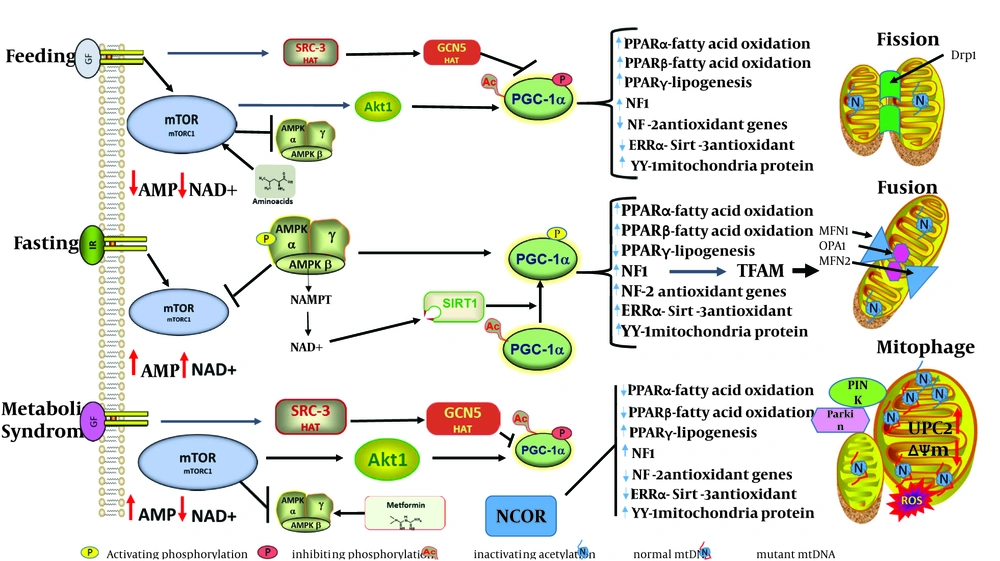
Obesity and over-nutrition, along with a lack of physical activity appear to be the initial environmental factors that initiate these metabolic abnormalities in genetically susceptible individuals. A central critical factor in the initiation of these metabolic anomalies is the inability of adipose, liver and muscle to metabolize excess fatty acids and carbohydrates, which leads to excess tissue lipid accumulation that induces a state of increased oxidative stress, which eventually leads to a persistent pro-inflammatory state that ultimately progresses to disease pathology. During caloric excess, one would expect the mitochondria to adapt by increasing fatty acid β-oxidation (FAO) and increasing the use of NADH by the electron transport – coupled with oxidative phosphorylation – to produce more ATP; however, a lack of physical activity reduces ATP necessity, thereby favoring a higher proto-motive force that can be dissipated by uncoupling protein to produce heat or lead to increased production of reactive oxygen species (ROS). Mitochondria are dynamic organelles that adapt to different nutrition conditions by increasing mitochondrial biogenesis and fission, which in turn increases ATP during low nutrient supply; during over-nutrition, mitochondria fuse and initiate mitophagy to control the number and quality of mitochondria (Figure 1).
Critical to the maintenance of mitochondrial function for adapting to different nutritional states is its ability to both increase the mitochondrial DNA transcription of 13 key essential proteins with the transcription of ~ 1500 nuclear genes and importation of these proteins to form multi-subunit complexes essential for proper mitochondrial function (2). This bi-genomic complex system is necessary for mitochondria to sense and adapt to different environmental nutrients, maintain mtDNA fidelity and through mitophagy, apoptosis or segregation remove dysfunctional mitochondria (3). Mitochondrial biogenesis, mtDNA replication and transcription of heavy and light mitochondrial strands are mediated by nuclear encoded mitochondrial transcription factor A (TFAM) (4). TFAM belongs to the high-mobility group (HMG) box domain proteins and has the ability to bind, bend and unwind mtDNA. Mitochondrial DNA are organized in protein-enriched nucleoids, with each nucleoid containing 1-2 mtDNA and one TRAM per 16.6 bp of the 16.5kbp mtDNA genome, thus identifying TFAM as the most abundant protein of nucleoids and possibly having the added function of protecting mtDNA from ROS damage (5, 6). Quantitative differences in the abundance of TFAM to mtDNA ratio differentially regulates mitochondrial replication and transcription, with low TFAM/mtDNA ratios favoring mtDNA replication, while high TFAM/mtDAN ratios favor transcription from the light (LSP) and heavy (HSP) promoters. TFAM importance in the maintenance of function has been elucidated in both knock-out and transgenic mouse models that revealed that a compensatory mechanism for mtDNA transcription exists in TFAM-KO mice; however, mtDNA depletion was evident due to dramatic respiratory enzyme deficiencies, strongly indicating that TFAM is essential for maintain the mtDNA copy number and thus essential for mitochondrial fission and mtDNA segregation (7).
With the realization of the central importance of TFAM in mitochondrial function, Mohammad Hashemi et al. in the April issue of Gene, Cell and Tissue (8), designed experiments to determine if there was an association of TFAM promoter regulation by methylation with susceptibility to metabolic syndrome in 300 male and female patients. Analysis of only one CpG region in the 5’ end of the region for DNA methylation led the authors to conclude that TFAM methylation was not associated with MeS. Unfortunately, the authors failed to measure either TFAM expression, mtDNA copy number, or other sites of methylation that may lead to reduced expression of TFAM and mtDNA copy number in this valuable patient group with metabolic syndrome (9, 10).
Several studies have analyzed nDNA and mtDNA methylation in metabolic disorders associated with metabolic syndrome. According to a recent study focused on DNA methylation of the Tfam promoter in peripheral leukocytes of high-school students, the ratio of promoter methylated DNA was inversely correlated with fasting plasma insulin, homeostasis model assessment index (HOMA) and obesity (11). A genome-wide analysis of DNA methylation and body-mass index revealed increased methylation of hypoxia inducing factor 3A (HIF3A) promoter to be associated with increased abdominal adipose and body mass index (BMI) (12). Additionally, increased PPARGC1A (PGC-1α) promoter methylation was correlated with HOMA-IR and plasma fasting insulin, while TFAM promoter CpG methylation was inversely correlated with fasting insulin in patients with NAFLD (13). In these patients, insulin resistance was correlated with a reduction in mtDNA copy number, even though the presence of mtDNA copy number in the liver of patients with NAFLD remains an unanswered question (14). These results suggest that mtDNA potentially plays an important role in alleviating some symptoms of MeS. Interestingly, in a transgenic mouse model of over-expressed human TFAM, there was an increase in mtDNA in all tissues without an increase in mitochondrial transcription or respiratory enzyme levels (15). In a heart-specific TFAM-KO mouse model showing dilated cardiomyopathy and conduction defects, overexpression of human TFAM restored mtDNA levels and rescued the cardiomyopathy phenotype (16).
It is presently unknown whether overexpression of TFAM can increase mtDNA levels in tissues affected by metabolic abnormalities associated with MeS. However, in patients with type 2 diabetes and insulin resistance (IR), the commonly prescribed drug, metformin, mimics low AMP levels, leading to the activation of AMP kinase and an amelioration of hyperglycemia. According to Figure 1, AMPK phosphorylation and the activation of PGC-1α will synergistically activate nuclear respiratory factor-2 (NRF-2) and consequently increase TFAM and mtDNA replication. It would be of interest to determine if patients with IR have an increased mtDNA copy number following treatment with this AMP kinase activator. A high caloric diet induces mitochondrial fusion and therefore increases mitochondrial size. In patients with NAFLD, a significant depletion of mtDNA alongside the presence of mega mitochondria with intra-mitochondrial crystalline inclusions are seen prior to progression to non-alcoholic steatohepatitis, suggesting that mitochondrial fusion may result in heteroplastic nucleoids, with one nucleoid having dysfunctional mtDNA (14). Unfortunately, very little is known about the control of mtDNA copy number per nucleoid or the number of nucleoids per mitochondria as it relates to TFAM function in these processes associated with MeS. However, the ability of over-expressed TFAM to stimulate oxidative metabolism in aged mice and restore non-mutated mtDNA replication in the cells of patients with Leber’s hereditary optic neuropathy (LHON) and Leigh syndrome indicate that mtDNA replication, mtDNA maintenance and nucleoid mtDNA copy number may be critical in the induction of fission and the segregation of dysfunctional mtDNA (15, 17). Indeed, recent evidence has shown that the inhibition of mitochondrial fission shifts the ratio of mutant mtDNA over normal mtDNA in a rhabdomyosarcoma cell line (18). Thus, the process of mitochondrial fusion and fission in the context of TFAM-induced mtDNA replication and possible nucleoid mtDNA copy number mediated segregation may play an important role in mitochondrial size, as may the number of mitochondria in tissues affected by metabolic abnormalities associated with metabolic syndrome.
It will be of utmost importance for Hashemi and others to use their valuable resources of patients with metabolic syndrome to examine nucleoid mtDNA copy number with TFAM expression in order to provide insight into mitochondrial size and number in their patients with MeS. The metabolic flexibility of mitochondria in response to different nutritional states is mediated by the integration of signal transduction events for modulating the expression of key nuclear and mitochondrial genes through transcription factors that are controlled by transcription factor co-activators and co-repressors (Figure 1). During caloric restriction or fasting, AMP levels increase, leading to AMPK activation, which phosphorylates and activates PGC-1α. An increase in Mitochondria NAD+ due to nutrient scarcity activates SIRT3, which de-acetylates and activates a number of mitochondrial proteins (superoxide dismutase, fatty acid oxidation long chain dehydrogenase, complex 1 protein NDUFA9), as well as the transcription histone acetylase, PGC-1α. PGC-1α co-activates a set of transcription factors involved in mitochondrial metabolism, PPARs, estrogen-related receptors, Yin-yang 1 and the all-important nuclear respiratory factor, which induces the transcription of mitochondrial transcription factor A (TFAM). Additionally, activation by deacetylation and phosphorylation of forkhead box O (FOXO) transcription factor results in the induction of genes involved in gluconeogenesis, fatty acid oxidation and NF1 controlled oxidative stress genes. FOXO also is a transcription factor in the regulation of mitochondria E3 ubiquitin ligase, which mediates mito-fusion MFN2 degradation and mitophagy. As cAMP levels rise and activate protein kinase A (PKA), PKA phosphorylates and inactivates Drp1, thus blocking mitochondrial fission.
During metabolic syndrome under conditions of caloric excess and lipid overload, mitochondrial fission is triggered, leading to mitochondrial uncoupling and the production of ROS. Decreased NAD+ reduces Sirt1 mediated activation of PGC-1α by deacetylation, while increased mitochondrial NADH levels increases acetyl-CoA, which serves as a substrate for SRC-3 and GCN5 acetylation and the inactivation of Sirt3 and PGC-1α. In a high-fat diet, the nuclear co-repressor 1 (NCOR1) is upregulated and represses the key transcription factors (NRF-1, NRF-2, ERRs and PPARs) that control mitochondrial activity, among them those that control expression of antioxidant genes. This results in increased mitochondrial ROS that can damage mtDNA and compromise mitochondrial function. Even with increased ROS and AMP levels to activate AMPK, PGC-1α is still inactivated by hyper-acetylation. Dysfunctional mitochondria, especially those with low membrane potential (ΔΨm) are recognized by the mitophagy process and eliminated. Unfortunately, under caloric excess, the balanced process of mtDNA replication and fission disconnects, leading to increasingly fragmented mitochondria with multiple copies of both functional and non-functional mtDNA (heteroplasmy).