1. Background
Analyzing genome variation is the most popular research in the field of genetic. This variation plays an important role in drug response and prediction of the disease (1). The Iranian population consists of around seventy million individuals consisting of people of many religions and ethnic backgrounds cemented by the Persian culture. Ethnic groups include Persians (51%), Azeris (24%), Gilaki and Mazandarani (8%), Kurds (7%), Arabs (3%), Baluchi (2%), Lurs (2%), Turkmens (2%) and others (1%) (2). These ethnic groups may have variations in their DNA sequences. The study of variations in DNA sequence is valuable when it is performed among individuals within the same population or among different populations. Tehran Lipid and Glucose Study (TLGS) is a prospective study of more than 15000 individuals (3-74 years) that live in the 13th district of Tehran Metropolitan. This study aims to develop population-based measures to alter the life-style and prevent the rising trend of non-communicable diseases. Furthermore, TLGS also aims to identify and tackle the risk factors for non-communicable diseases in a representative sample of individuals residing in Tehran, who were recruited by a stratified cluster sampling method (3, 4). Metabolic syndrome is a combination of medical disorders that increases the risk of developing cardiovascular disease and diabetes (5). The prevalence of the metabolic syndrome is 32% in adults (6) and 10% in adolescents (7). In the present study, we aimed to determine the allele frequency of five microsatellite markers on chromosome 12 and 16, and to observe possible differences in the genetic patterns among several ethnic groups. In addition, this study investigated genetic variation between people with and without the metabolic syndrome.
2. Objectives
This article reports primary results in the form of allele frequency distributions and summary statistics of five different autosomal STR loci.
3. Materials and Methods
Population: based on the frequency of metabolic syndrome, a total of 563 individuals aged 3-87 were randomly selected from the TLGS for analyzing the allele frequency of four microsatellite markers on chromosome eight. All subjects answered a questionnaire covering data on demographic factors, smoking habits and other relevant information. Written informed consent was obtained from each subject. The research council of the Endocrine Research Center of the Shahid Beheshti University of Medical Sciences (M.C) approved this study.
3.1. Ethnic Groups
Four ethnic groups were included: Persian (68%), Turk (18.3%), Mazani/Gilaki (8%) and Kurd/Lur (6.2%). To simplify the analysis, the Mazani and Gilaki (people of northern Iran) and the Lur and Kurd (people of western Iran) were combined.
Metabolic syndrome was defined as a cluster of metabolic risk factors for cardiovascular diseases and type 2 diabetes mellitus. Metabolic syndrome X consists of the following complications, excess abdominal fat, atherogenic dyslipidemia, hypertension, hyperglycemia, insulin resistance, a proinflammatory state and a prothrombotic (thrombosis) state (8).
The following phenotypic measurements were obtained for each subject: body mass, body mass index (BMI), height and blood pressure. Blood samples were collected in EDTA containing tubes and serum in tubes without any anticoagulant. After centrifugation for 10 minutes at 3000 rpm, sera were separated and stored at -70°C in 1.5 mL aliquots. Serum glucose, total cholesterol, high-density lipoprotein-cholesterol (HDL-C) and triglyceride levels were measured immediately from fresh sera as described previously (9). Serum HDL-C levels were measured after precipitation of Apo B containing lipoproteins with dextran-magnesium sulfate (10). Low-density lipoprotein-cholesterol (LDL-C) and very low-density lipoprotein (VLDL) concentrations in samples with serum triglyceride levels < 400 mg/dL were calculated using Friedewald’s equation, and one fifth of triglyceride level, respectively (11). Coefficients of variation (CV) for total cholesterol, HDL-C and triglyceride measurements were below 5%.
3.2. DNA Analysis
When genomic DNA was extracted by the proteinase K and salting out standard method, buffy coats were separated from the non coagulated blood samples and stored at -70°C until processing (12). The GeneAmp PCR System 9700 (ABI USA) was used to simultaneously amplify the five STRs loci including D12S1632, D12S329, D12S96, D16S3096 and D16S2624. The characteristics of STRs loci are presented in Table 1. Four out of five have dinucleotide repeats and one has a tetranucleotide repeat. Amplification was performed using 100 ng of total genomic DNA in a final volume of 25 L containing 5 pmol of each primer and gold mix of Taq polymerase (ABI USA).
The amplification conditions were as follows: 95˚C for 11 minutes, followed by 30 cycles of 30 seconds at 94˚C, 60 seconds at 55˚C and 40 seconds at 72˚C, and ending with a single 30-minute extension step at 72˚C. Electrophoresis of the amplification products was performed on an ABI 3100 Genetic Analyzer (Applied Biosystems Co.). The raw data were analyzed by the ABI Data Collection Software and GeneMapper 3.2 (Applied Biosystems). For quality control laboratory internal control standards were used.
| Locus | Location | Repeats Unit | Sequence of Primers | Polymorphic Region |
|---|---|---|---|---|
| D12S96 | 12q13.13 | [CA]n | CCAGTTCAAACCAGTGACCT Labeled with (PET) | 201-227 |
| TCCATCCTTGTGGGCA | ||||
| D12S1632 | 12q13.2 | [TG]n | GCCTAATCAAGATGTCACCA Labeled with (VIC) | 208-230 |
| GCTAGGGAGCCAATTCA | ||||
| D12S329 | 12q14.2 | [GT]n | AAGCAATCAGCCAGCCCT Labeled with (NED) | 143-171 |
| TGTCAGAACCTAACAACCCAGAAAG | ||||
| D16S2624 | 16q22.3 | [ATCT]n | TGAGGCAATTTGTTACAGAGC Labeled with (6-FAM) | 130-148 |
| TAATGTACCTGGTACCAAAAACA | ||||
| D16S3096 | 16q23.1 | [GT]n | GATCTGGCTTACGATGATTTCTAAC Labeled with (PET) | 199-229 |
| CCGTGATGATGTCTGCAAC |
3.3. Statistical Analysis
Explanatory statistics were used for population characteristics and data are shown as mean ± standard deviation for normally distributed variables and as percentages for categorical variables. Differences between ethnic groups were evaluated by Student's t-test for normally distributed data. The distribution of the triglycerides was skewed, and a comparison was performed using Mann–Whitney’s U-test. Analysis of categorical variables was performed by Chi-square and Fisher’s exact tests for contingency tables. Allele frequency and polymorphic information content (PIC) values were computed by the PowerMarker software (13, 14). Deviation from Hardy-Weinberg equilibrium, as well as observed and expected heterozygosity, were calculated using the GenePop software Version 3.4 (15). The Excel PowerStats spreadsheet from promega (16) was used to calculate discrimination power, matching probability, power of discrimination, power of exclusion and paternity index.
4. Results
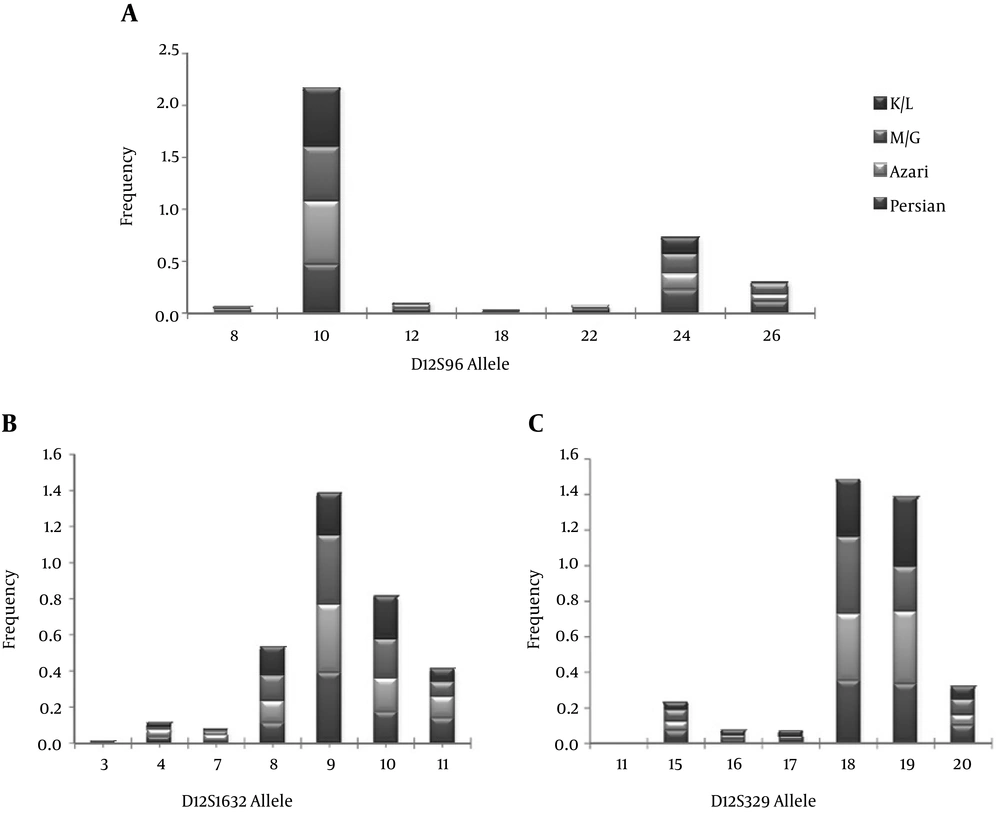
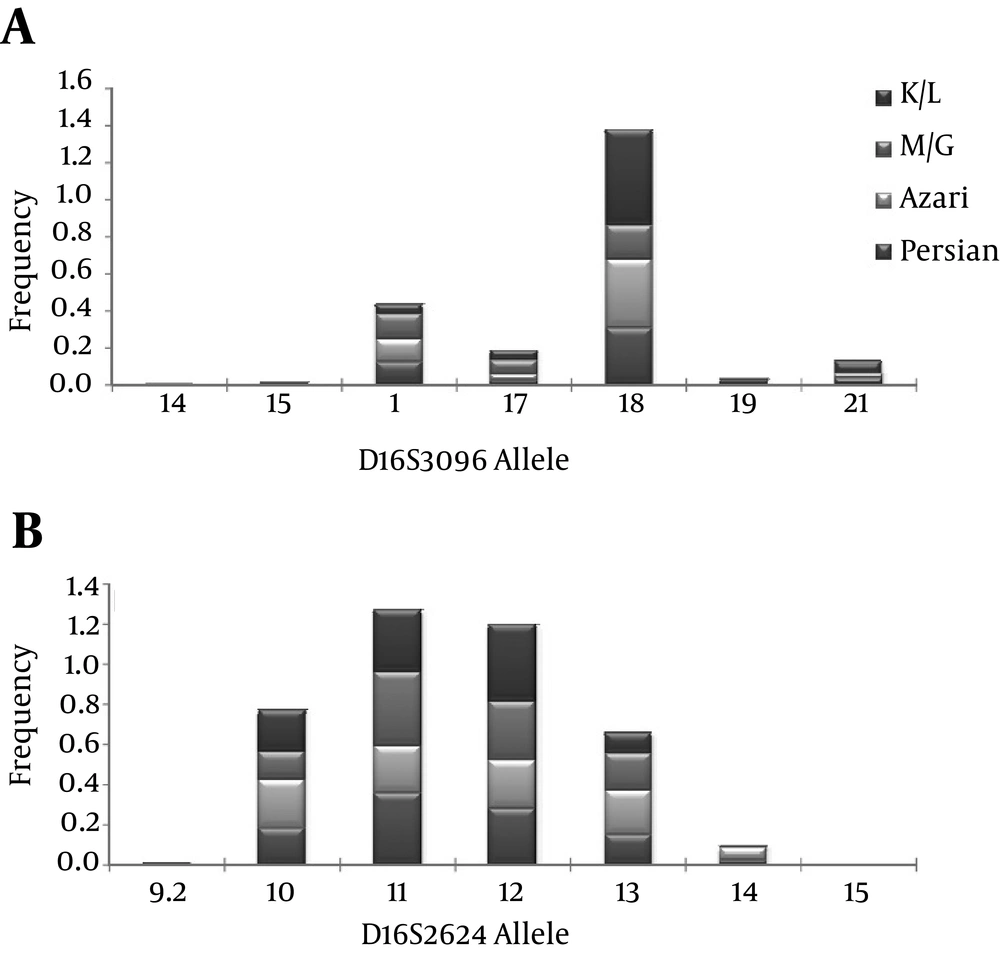
The demographic and biochemical parameters of 563 participants consisting of 270 men and 293 women with the mean age of 36 ± 19 are shown in Table 2. There were no statistically significant differences between ethnic groups in biochemical characteristics related to the metabolic syndrome. The allele frequencies for the five STRs loci in 563 unrelated Tehranian samples are presented in Table 3. The most polymorphic marker is D16S2624, this marker has a wide range of size with seven different alleles. Sample populations were observed to be in Hardy-Weinberg Equilibrium (HWE) for all analyzed markers (P < 0.05), except for D12S1632 and D12S329. Some factors such as: matching probability, power of discrimination, power of exclusion and paternity index were calculated for this population. Allele frequency distribution in these five microsatellites in four ethnic groups are presented in Figures 1-2 the details of the allele frequencies, matching probability, power of discrimination, power of exclusion and paternity index in each ethnic group are presented in Table 4.
For the D12S96 microsatellite, a total of 11 alleles were observed in the 563 subjects. These were named 201-227 (PCR product length), which correspond to 6-32 (CA)n repeats, respectively. The microsatellite length was used to subdivide samples into two groups according their size: [short (≤ 213), long (> 217)]; [short (≤ 207), medium (207-221) and long (≥ 221)] for case-control analysis. For the D12S1632 microsatellite, a total of 10 alleles were observed in 563 subjects. These were named 208-230 (PCR product length), which corresponded to 4-15 (TG)n repeats, respectively. The microsatellite length was used to subdivide samples into two groups according their size: [short (≤ 220), long (> 220)]; [short (≤ 216), medium (218-222), long (≥ 224)] for case-control analysis. For the D12S329 microsatellite, a total of 10 alleles were observed in 563 subjects. These were named 143-171 (PCR product length), which corresponded to 11-25 (GT)n repeats, respectively. The microsatellite length was used to subdivide samples into two groups according their size: [short (≤ 157), long (> 159)]; [short (≤ 153), medium (155-161), long (≥ 163)] for case-control analysis. For the D16S2624 microsatellite, a total of six alleles were observed in 563 subjects. These were named 130-148 (PCR product length), which corresponded to 9.2-14 (ATCT)n repeats, respectively. The microsatellite length was used to subdivide samples into two groups according their size: [short (≤ 136), long (> 140)]; [short (≤ 136), medium (136-140), long (≥ 140)] for case-control analysis. Finally, for the D16S3096 microsatellite, a total of 17 alleles were observed in 563 subjects. These were named 199-229 (PCR product length), which corresponded to 14-29 (GT)n repeats, respectively. The microsatellite length was used to subdivide samples into two groups according their size: [short (≤ 215), long (> 216)]; [short (≤ 209), medium (209-219), long (≥ 219)] for case-control analysis. The allele frequencies of the control and the metabolic syndrome groups were compared for the four microsatellites subdivided in three groups (short, medium and long). In the D12S329, the frequency of long alleles in subjects with metabolic syndrome was significantly higher than the controls (P < 0.05) (Table 5). In Table 6 the allele frequencies of the four different ethnic groups were compared for the five microsatellites in two subdivided groups (short and long) and were significantly different.
| Characteristic | Total (n = 563) | Persian (n = 380) | Turk (n = 103) | Mazani/Gilaki (n = 45) | Kurd/Lur (n = 35) |
|---|---|---|---|---|---|
| Metabolic syndrome | 130 (28) | 82 (21.5) | 23 (22.3) | 11 (24.5) | 14 (40) |
| Age, y | 36 ± 19 | 35 ± 19 | 37 ± 19 | 38 ± 18 | 39 ± 20 |
| Sex, females, % | 52 | 51.3 | 49.5 | 53.3 | 68.6 |
| BMI, kg/m2 | |||||
| Women | 25 ± 6 | 26 ± 6 | 26 ± 5 | 27 ± 7 | 26 ± 6 |
| Men | 25 ± 5 | 25 ± 5 | 25 ± 5 | 25 ± 5 | 29 ± 5 |
| Family history of diabetes, % | 7.3 | 8.0 | 3.9 | 8.9 | 8.8 |
| Components of metabolic syndrome | |||||
| Waist circumference, cm | |||||
| Women | 83 ± 15 | 83 ± 15 | 84 ± 16 | 86 ± 15 | 84 ± 16 |
| Men | 90 ± 15 | 89 ± 15 | 90 ± 13 | 88 ± 14 | 100 ± 12 |
| Fasting plasma glucose, mg/dL | 95 ± 29 | 95 ± 30 | 92 ± 19 | 100 ± 37 | 93 ± 19 |
| Elevated blood pressure, mmHg | |||||
| Systolic | 113 ± 29 | 113 ± 20 | 113 ± 19 | 112 ± 20 | 117 ± 25 |
| Diastolic | 71 ± 11 | 70 ± 10 | 69 ± 11 | 70 ± 10 | 74 ± 13 |
| Serum triglycerides, mg/dL | 140 ± 87 | 137 ± 79 | 143 ± 90 | 154 ± 130 | 148 ± 83 |
| HDL cholesterol, mg/dL | |||||
| Women | 47 ± 1 | 47 ± 11 | 46 ± 11 | 48 ± 12 | 47 ± 12 |
| Men | 41 ± 9 | 41 ± 9 | 40 ± 9 | 473 ± 12 | 35 ± 7 |
aData are presented as Means ± SD, No. (%) or %.
| Repeat | D16S2624 | D16S3096 | D12S1632 | D12S329 | D12S96 |
|---|---|---|---|---|---|
| 2 | 0.0009 | ||||
| 3 | 0.0009 | ||||
| 4 | 0.0333 | ||||
| 6 | 0.0026 | ||||
| 7 | 0.0144 | ||||
| 8 | 0.1180 | 0.0212 | |||
| 9 | 0.3793 | ||||
| 9.2 | 0.0111 | ||||
| 10 | 0.1908 | 0.1811 | 0.5053 | ||
| 11 | 0.3228 | 0.1252 | 0.0017 | ||
| 12 | 0.2785 | 0.1144 | 0.0362 | ||
| 13 | 0.1695 | 0.0162 | |||
| 14 | 0.0256 | 0.0045 | 0.0072 | ||
| 15 | 0.0017 | 0.0009 | 0.0090 | 0.0693 | |
| 16 | 0.1203 | 0.0197 | |||
| 17 | 0.0305 | 0.0094 | |||
| 18 | 0.3142 | 0.3622 | 0.0097 | ||
| 19 | 0.0171 | 0.3416 | |||
| 20 | 0.0018 | 0.0873 | |||
| 21 | 0.0296 | 0.0771 | |||
| 22 | 0.0180 | 0.0291 | 0.0362 | ||
| 22.1 | 0.0036 | ||||
| 23 | 0.0637 | ||||
| 24 | 0.1059 | 0.2046 | |||
| 25 | 0.2621 | 0.0026 | |||
| 26 | 0.0108 | 0.0935 | |||
| 27 | 0.0108 | ||||
| 28 | 0.0036 | 0.0309 | |||
| 29 | 0.0027 | ||||
| 30 | 0.0556 | ||||
| 32 | 0.0044 | ||||
| PIC | 0.7100 | 0.7750 | 0.7525 | 0.6918 | 0.6558 |
| Ho | 0.8395 | 0.8072 | 0.7900 | 0.7736 | 0.6474 |
| He | 0.7523 | 0.7986 | 0.7740 | 0.7370 | 0.6859 |
| MP | 0.1174 | 0.0728 | 0.0778 | 0.1170 | 0.1393 |
| PD | 0.8826 | 0.9272 | 0.9222 | 0.8830 | 0.8607 |
| PE | 0.6584 | 0.6341 | 0.5693 | 0.5365 | 0.3539 |
| PI | 2.9646 | 2.7574 | 2.3125 | 2.1314 | 1.4246 |
| P | 0.0060 | 0.0000 | 0.8800 | 0.9850 | 0.0000 |
aAbbreviations: Hobs, Observed heterozygosity; Hexp, expected heterozygosity; MP, matching probability; PIC, Polymorphic information content; P, probability value of exact tests of Hardy-Weinberg disequilibrium; PD, discrimination power; PE, power of exclusion; PI, paternity index.
| Persian | Azeri | Mazani/Gilaki | Kurd/Lur | |
|---|---|---|---|---|
| D12S96 | ||||
| MP | 0.133 | 0.202 | 0.152 | 0.189 |
| PD | 0.867 | 0.798 | 0.848 | 0.811 |
| PIC | 0.675 | 0.573 | 0.640 | 0.579 |
| PE | 0.354 | 0.270 | 0.422 | 0.412 |
| PI | 1.43 | 1.20 | 1.64 | 1.61 |
| Allele Frequencies | ||||
| Homozygotes, % | 35.1 | 41.7 | 30.4 | 31.0 |
| Heterozygotes, % | 64.9 | 58.3 | 69.6 | 69.0 |
| Total Alleles | 724 | 206 | 92 | 58 |
| D12S329 | ||||
| MP | 0.116 | 0.149 | 0.105 | 0.157 |
| PD | 0.884 | 0.851 | 0.895 | 0.843 |
| PIC | 0.703 | 0.633 | 0.692 | 0.681 |
| PE | 0.551 | 0.453 | 0.529 | 0.643 |
| PI | 2.21 | 1.76 | 2.09 | 2.83 |
| Allele Frequencies | ||||
| Homozygotes, % | 22.7 | 28.4 | 23.9 | 17.6 |
| Heterozygotes, % | 77.3 | 71.6 | 76.1 | 82.4 |
| Total Alleles | 750 | 204 | 92 | 68 |
| D12S1632 | ||||
| MP | 0.084 | 0.079 | 0.116 | 0.090 |
| PD | 0.916 | 0.921 | 0.884 | 0.910 |
| PIC | 0.743 | 0.761 | 0.738 | 0.791 |
| PE | 0.586 | 0.532 | 0.500 | 0.588 |
| PI | 2.41 | 2.11 | 1.95 | 2.43 |
| Allele Frequencies | ||||
| Homozygotes, % | 20.7 | 23.7 | 25.6 | 20.6 |
| Heterozygotes, % | 79.3 | 76.3 | 74.4 | 79.4 |
| Total Alleles | 714 | 194 | 86 | 68 |
| D16S2624 | ||||
| MP | 0.126 | 0.108 | 0.145 | 0.168 |
| PD | 0.874 | 0.892 | 0.855 | 0.832 |
| PIC | 0.695 | 0.739 | 0.674 | 0.651 |
| PE | 0.643 | 0.779 | 0.529 | 0.588 |
| PI | 2.83 | 4.64 | 2.09 | 2.43 |
| Allele Frequencies | ||||
| Homozygotes, % | 17.7 | 10.8 | 23.9 | 20.6 |
| Heterozygotes, % | 82.3 | 89.2 | 76.1 | 79.4 |
| Total Alleles | 758 | 204 | 92 | 68 |
| D16S3096 | ||||
| MP | 0.076 | 0.083 | 0.111 | 0.130 |
| PD | 0.924 | 0.917 | 0.889 | 0.870 |
| PIC | 0.774 | 0.753 | 0.719 | 0.666 |
| PE | 0.670 | 0.531 | 0.591 | 0.510 |
| PI | 3.08 | 2.10 | 2.44 | 2.00 |
| Allele Frequencies | ||||
| Homozygotes,% | 16.2 | 23.8 | 20.5 | 25.0 |
| Heterozygotes, % | 83.8 | 76.2 | 79.5 | 75.0 |
| Total Alleles | 714 | 202 | 88 | 56 |
aAbbreviations: MP, matching probability; PIC, Polymorphic information content; PD, discrimination power; PD, power of discrimination; PE, power of exclusion; PI, paternity index.
| Non Metabolic Syndrome | Metabolic Syndrome | |
|---|---|---|
| D12S96 | ||
| Short allele | 0.5581 (480)a | 0.5977 (159) |
| Medium allele | 0.2465 (212) | 0.2481 (66) |
| Long allele | 0.1953 (168) | 0.1541 (41) |
| D12S329 | ||
| Short allele | 0.0887 (79) | 0.0919 (25) |
| Medium allele | 0.8146 (725)b | 0.7610 (207) |
| Long allele | 0.0966 (86) | 0.1470 (40) |
| D12S1632 | ||
| Short allele | 0.0305 (26) | 0.0465 (12) |
| Medium allele | 0.5176 (441) | 0.4961 (128) |
| Long allele | 0.4518 (385) | 0.4573 (118) |
| D16S3096 | ||
| Short allele | 0.1662 (140) | 0.1893 (50) |
| Medium allele | 0.2220 (187) | 0.2234 (59) |
| Long allele | 0.6116 (515) | 0.5871 (155) |
| D16S2624 | ||
| Short allele | 0.5339 (441) | 0.5233 (135) |
| Medium allele | 0.2772 (229) | 0.2752 (71) |
| Long allele | 0.1889 (156) | 0.2016 (52) |
aNumber in parenthesis denote number of subjects
bLong allele frequency vs. medium allele frequency, significance at P < 0.001 by the Chi-square test.
| Marker | Persian | Azeri | Mazani/Gilaki | Kurd/Lur |
|---|---|---|---|---|
| D12S96a | ||||
| Short allele | 0.5360 (387)b | 0.6747 (139) | 0.5888 (53) | 0.5689 (33) |
| Long allele | 0.4639 (335) | 0.3252 (67) | 0.4111 (37) | 0.4310 (25) |
| D12S329 | ||||
| Short allele | 0.4558 (341) | 0.4460 (91) | 0.5333 (48) | 0.4264 (29) |
| Long allele | 0.5441 (407) | 0.5539 (113) | 0.4666 (42) | 0.5735 (39) |
| D12S1632 | ||||
| Short allele | 0.5407 (385) | 0.5721 (111)c | 0.5714 (48) | 0.4411 (30) |
| Long allele | 0.4592 (327) | 0.4278 (83) | 0.4285 (36) | 0.5588 (38) |
| D16S3096 | ||||
| Short allele | 0.4747 (338)d | 0.5396 (109)e | 0.4069 (35)f | 0.6250 (35) |
| Long allele | 0.5252 (374) | 0.4603 (93) | 0.5930 (51) | 0.3750 (21) |
| D16S2624 | ||||
| Short allele | 0.5489 (415)g | 0.4754 (97) | 0.5222 (47) | 0.5147 (35) |
| Long allele | 0.4510 (341) | 0.5245 (107) | 0.4777 (43) | 0.4852 (33) |
aNumber in parenthesis denote number of subjects.
bPersian allele frequency vs. Azeri allele frequency, significance at P < 0.001 by the Chi-square test.
cAzeri allele frequency vs. Kurd/Lur allele frequency, significance at P < 0.001 by the Chi-square test.
dPersian allele frequency vs. Kurd/Lur allele frequency, significance at P < 0.001 by the Chi-square test.
eAzeri allele frequency vs. Mazani/Gilaki allele frequency, significance at P < 0.001 by the Chi-square test.
fKurd/Lur allele frequency vs. Mazani/Gilaki allele frequency, significance at P < 0.001 by the Chi-square test.
gPersian allele frequency vs. Azeri frequency, significance at P < 0.001 by the Chi-square test.
5. Discussion
This study is the first allele frequency report related to chromosomes 12 and 16 in the Iranian population. Based on our knowledge there has been no allele frequency data for our selected microsatellite on chromosome 12. To confirm the new allele in D16S2624, the ALFERED database was accessed. Furthermore, in subjects with metabolic syndrome, the long alleles were significantly more frequent in D12S329 (P < 0.05). Between the different ethnic groups there were some differences in short and long allele frequencies. D16S2624 is a tetra nucleotide repeat marker but in this population one new allele (15.2) was seen. The most heterozygote marker in the total population and in different ethnic groups was D16S2624. The power of discrimination ranged from a minimum of 0.798 for D12S96 locus in the Azeri group to a maximum of 0.924 for D16S3096 locus in the Persian group. In the Iranian population, the distribution of the analyzed loci alleles was not previously studied. The present dataset will add to the reference database and will be helpful in population genetics and diversity studies. Differences between medium and long allele frequencies in D12S329 of subjects with metabolic syndrome in comparison with controls may be a sign of association between this region and the presence of the metabolic syndrome. Further analysis with more microsatellites in this region can lead us to the genetic cause of metabolic syndrome. Ethnic groups have some variation in allele frequency but the sample size was not big enough to reach a comprehensive conclusion. In the future the most important markers in this population have to be checked in order to improve knowledge regarding the genetic pattern of the Iranian population.