1. Context
The impact of cancer on humanity is huge and a great deal of research is going on worldwide to find novel therapeutic targets. Telomerase is one such target. Increased telomerase activity provides cancer cells with unlimited proliferative potential and is one of the hallmarks of cancer. This article provides a basic understanding of telomere and telomerase in cancer and summarizes some potential therapeutic approaches used for strategic targeting of the telomerase enzyme (1).
2. Evidence Acquisition
2.1. Telomere Structure and Function
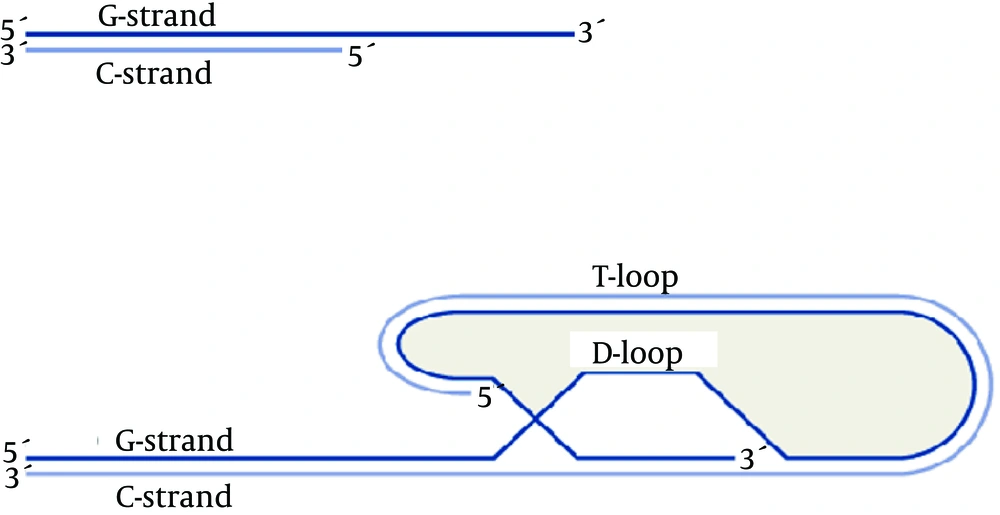
A telomere consists of two C-rich and G-rich strands of nucleotides; end of G-rich end of C-rich nucleotides. In human beings and other vertebrates, telomeres are located on chromosome ends and each one contains thousands of tandem repeat sequences of 5’(TTAGGG)3’(2). However, other species have different sequences on their chromosome ends; for example, Tetrahymena has 5’(TTGGGG)3’n and Schizosaccharomyces pombe has 5’GGTTAC(A)(C)(G0-6)3’. In humans, these repetitive sequences are 15 - 20 kb in length at birth and about 8 - 10 kb in adults. It is worth noting that these repetitive sequences have different lengths between people, cells and even between various chromosomes. The 3’ end of every chromosome is single-stranded, does not have a complementary strand, and is about 200 nucleotides long (3). Both single stranded 3’ ends of each chromosome loops back, anneals to the double-stranded part, and forms a T loop with a part of the double-stranded section. After this looping back and T loop formation, the single stranded DNA forms a hydrogen bond with its repetitive complementary sequence on the 5’ end (which is double-stranded) and thus, through the formation of a displacement loop, the other strand of the DNA becomes double-stranded. That is why this loop is called the displacement loop or D loop (Figure 1) (4).
T-loop and D-loop in the Telomere Structure (5)
Telomeres protect chromosome ends from degeneration, DNA repair activities, and end-to-end fusion. Uncapped telomeres activate responses to DNA damage and, as a result, due to the end-to-end fusion that follows these responses, cell aging, apoptosis, and chromosomal instability increase. The presence of telomeres, because of their position, suppresses adjacent genes and prevents their expression. Shortening of telomeres can cause these silent genes to express themselves (6). Moreover, it seems that telomeres play an important role in the formation of bouquets at the start of meiosis and, therefore, telomere inefficiency can lead to death of germ cells (7).
2.2. Telomere Binding Proteins
The repetitive and highly protected sequence of human telomeres led research towards finding factors that specifically bind to these repetitive sequences. TTAGGG-Repeat binding Factor 1 (TRF1), due to its specific binding to the double-stranded part of TTAGGG repeats in Hela cells, was the first factor to be identified. Since then, five other binding proteins have been identified, including TIN2, RAP1, POT1, TRF2 and TPP1. These six proteins together form the shelterin complex that specifically covers telomeric DNA and is considered an essential factor for maintaining the integrity of chromosomes and for preventing genomic instability (8). In addition to the proteins in the shelterin complex, considered the main proteins that bind to telomeres, DNA repair proteins including Ku complex, BLM /WRN, ATM, XPF/ERCC1, MRN complex (MRE11, RAD50, and NBS1), RAD51D and RAD 54, also play a role in the preservation and survival of telomeres. Mutation or absence of such genes, can lead to the shortening and end-to- end fusion of telomeres and to the generation of premature aging phenotypes (9).
2.3. The Reason Behind the Gradual Shortening of Telomeres
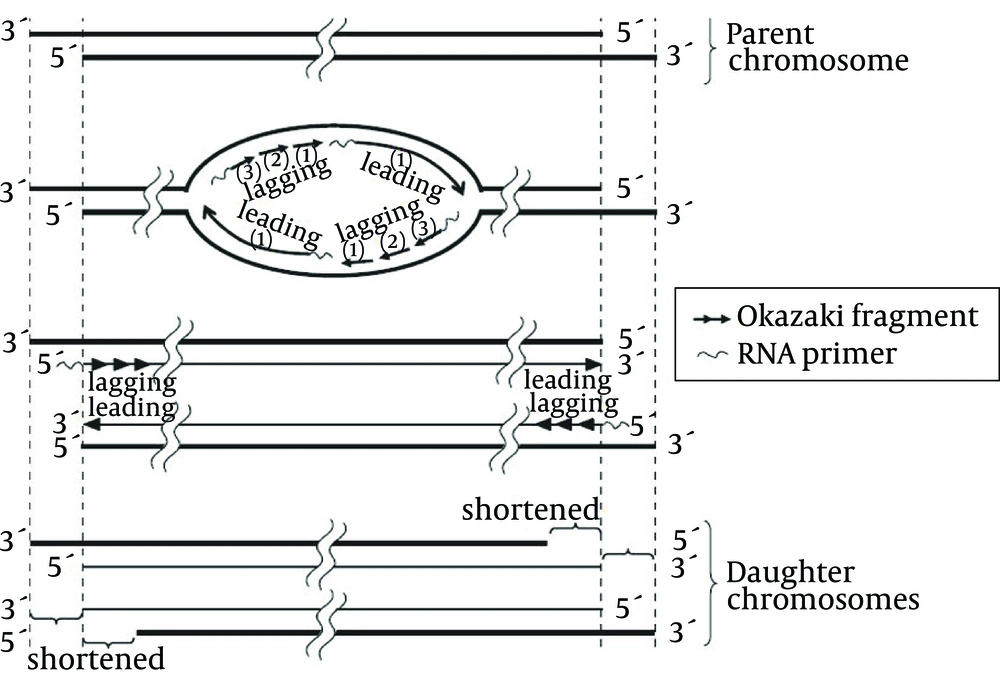
Until the early 1960s, it was thought that normal human cells cultured in the lab would indefinitely divide if suitable conditions were maintained. In 1961, Hayflick showed that the number of times normal human cells divide is limited; pulmonary fibroblasts divided six times, heart cells 26, kidney cells 40, and skin cells 43 times. Therefore, this phenomenon was called the Hayflick limit (10). In 1971 and 1972, Olovnikov and Watson reported the mechanism of this limit and called it the end-replication problem. They claimed that DNA replicates only in the 5’ to 3’ direction and needs RNA primers and complementary strands for this replication and, hence, after deletion of RNA primers on the 5’ end of the lagging strand, no other DNA can displace replicating the region any longer. In their reports, they said the lagging strand was responsible for telomere shortening and called this problem the lagging strand problem (11). However, considering the 3’ overhang structure and the inability of the leading strand in synthesizing the 3’ overhang, the leading strand problem was finally introduced as the chromosome end replication problem (Figure 2) (12).
Incomplete replication of the leading strand due to the formation of 3’overhang, as well as that of the lagging strand due to RNA primer as a prerequisite for DNA synthesis, are responsible for telomere shortening (13).
If organisms cannot solve this replicative problem they cannot pass the genetic charge completely from generation to generation. As a consequence, all the species should have, at least at the level of germinal cells, a mechanism that prevents the incomplete replication of the genomes (14).
2.4. Telomerase Structure and Function
Somatic cells cultured in the lab can divide only a limited number of times and the reason for this may be the lack, or the presence of low levels, of telomerase and the gradual shortening of telomere length in successive cell divisions. On the contrary, cells like male germ line stem cells, activated lymphocytes, hematopoietic progenitor cells and basal keratinocytes exhibit considerable division capability due to the presence of the ribonucleoprotein enzyme telomerase (the only enzyme that is responsible for the elongation of telomeres in cells) (15, 16).
Human telomerase is a ribonucleoprotein enzyme that consists of a catalytic component and an RNA component. The catalytic component has 1132 amino acids, a molecular weight of 127 kd, and is called the human telomerase reverse transcriptase (hTERT) (17).
The telomerase enzyme also has an RNA component as a template that is called human RNA telomerase or the RNA component of the telomerase (hTR: human Telomerase RNA or TERC: Telomerase RNA Component) (18). The 3’end of telomeres (3’overhang) is single-stranded and can bind to the telomerase RNA template succession. Telomerase is a reverse transcriptase enzyme that uses an RNA molecule as a template to synthesize DNA. In other words, telomerase is an RNA dependent DNA polymerase. Telomere elongation can be considered as the best-known function of telomerase, and cell replication capacity increases due to telomerase activity and cell division sometimes continues indefinitely.
2.5. Method of Measurement of Telomerase Activity
The telomeric repeat amplification protocol (TRAP) assay has been the standard assay in the detection of telomerase activity and many variations of this technique have been reported. This method is divided into three main steps that consist of the extension by telomerase, amplification by polymerase chain reaction (PCR), and the detection of telomerase products. In the extension step, telomerase in the cell extract adds telomeric repeat to the substrate (TS), a non-telomeric oligonucleotide. The extension products are amplified about a million folds by the PCR using specific primers, and in most cases the amplified products are analyzed by electrophoresis (13).
2.6. Telomerase in Normal, Cancer and Stem Cells
Telomerase activity has not been detected in most normal human somatic cells, yet lymphocytes and most, but not all, stem/progenitor cells in self-renewal tissues can express telomerase upon mitogenic stimulation (19). These cells have a long lifetime and, when their division is stopped or differentiated, their telomerase activity disappears.
Normal human somatic cells, even those in which telomerase activity has been detected, never become immortal unless their tumor-suppressor genes are suppressed or their oncogenes become active (20, 21).
Human mesenchymal stem cells, despite having a very low level of telomerase activity, have longer telomeres than somatic cells. In addition, these cells do not possess characteristics such as the presence of positively-charged poly-L-lysine (PLL) bodies observed in Alternative lengthening of telomerescells (ALT) , and it is said that they may have their own specific and different chromosome survival mechanisms (22).
Normal cells must overcome the two stages of mortality mechanism referred to as the M1 and M2 in order to evade cell aging and to become immortal (23). In normal cells that lack telomerase, cell division stops at the M1 stage due to the shortening of telomeres at each cell division stage and because of the consequent identification of uncapped telomeres as damaged DNA by P53 and p16/p Rb, and cellular senescence occurs. With the inactivation of these tumor suppressor genes, cells can pass the M1 stage and continue dividing and their lifetime increases. However, this cell division is stopped again at the crisis or the M2 stage. At this stage, many telomeres are critically shortened, end-end fusions occur, and cells stop dividing, and, eventually, apoptosis results (that is independent of p53) (24). The escape from M2 (the crisis stage) is a rare phenomenon in human cells and, therefore, it can be said that senescence and the crisis stage are two anticancer mechanisms in species like humans that have long lifetimes. While the main mechanism in most cancer cells for evading the crisis stage and for becoming immortal is telomerase gene expression, there is at least one other mechanism called alternative lengthening of telomeres (ALT) in some cancer cells for the preservation and survival of telomeres that allows cells to evade the crisis stage through DNA recombination (25-27).
3. Results
3.1. Telomerase Inhibition
The main characteristic of cancer cells is their capacity to replicate without limits, which is achieved by telomerase-mediated telomere maintenance in the majority of advanced tumors (28). Therefore, telomerase inhibitors, which destroy the capacity of telomerase-containing cancer cells to replicate, can be applied as selective anticancer treatment (29). Since telomerase levels in normal cells are very low, using telomerase inhibitors cannot significantly affect these cells (30). However, in addition to cancer cells, stem cells and germ line cells in the human body also have high levels of telomerase. The answer to the question of whether application of telomerase inhibitors will damage these two classes of cells is that, although telomerase levels in these two cell classes are high (as they are in cancer cells), their telomere length is different from and longer than that of cancer cells and, therefore, before they can be damaged, tumor cells experience apoptosis (20, 31).
Telomerase regulation is complex and has important consequences not just for telomere length, but also for the development of disease. Understanding how telomerase is regulated will also lead to improved targeting of telomerase as a therapy for genetic diseases and cancer (32).
There are two general strategies to inhibit telomerase activity in cancer cells. One is a direct mechanism, i.e. using compounds that directly cause telomerase inhibition by inhibiting the activity of the catalytic subunit (hTERT), the RNA template (hTR) or the telomere structure. Indirect strategies involve blocking telomerase access to telomeres by using G-quadruplex stabilizers, or by inhibiting binding of telomerase-associated proteins leading to telomere uncapping and cell apoptosis (1).
3.2. Targeting Human Telomerase Reverse Transcriptase and Human Telomerase RNA
By the use of nucleic acid–based biological sequences, such as small interfering RNA (siRNA), siRNA expression vectors, ribozymes and antisense oligonucleotides, telomerase catalytic subunit hTERT can be inhibited (1).
Human Telomerase Reverse Transcriptase has recently been shown to be affected by MicroRNAs (32). The MicroRNAs, also known as miRNAs or miRs, are noncoding, single stranded RNAs that have recently been shown to play important roles in many biological processes such as development, differentiation, apoptosis and proliferation (33). More than 1000 miRNAs are predicted to work in human biology in a complex regulation network, and several studies have shown that many miRNAs are deregulated in cancers (33-35). Recently, transfected thyroid carcinoma cell lines with miR-138 precursor molecules, showed reduced hTERT expression, and shorter life spans. Additionally, the miR-375 as a tumor suppressor miRNA has been shown to regulate cell cycle and telomerase activity. Furthermore, miR-375 inhibits nuclear translocation of TERT, thereby contributes to reduce telomerase activity (36).
Ribozymes are small RNA molecules that have specific endoribonuclease activity. They consist of a catalytic core, flanked by anti-sense sequences that function in the recognition of the target site (37). Hayashidani et al. demonstrated the inhibition of telomerase activity by ribozymes in pancreatic cell lines (38).
Among small molecule inhibitors, BIBR1532 (2-[E)-3-naphtalen-2-yl-but-2-enoylylamino]- benzoic acid), a synthetic non-competitive inhibitor of hTERT has been developed thus far. It directly interacts with hTERT component of telomerase. Results from various studies have demonstrated a dose-dependent inhibition of telomerase with high concentration of BIBR1532 with no significant effect on normal cells. Good results have been observed in preclinical studies on breast, prostate and fibrosarcoma cancer cell lines (1).
Telomerase is dependent on its (hTR) RNA sequence in building repetitive sequences of telomere, and because its RNA sequence is known, targeting this region of the enzyme by small oligonucleotides that can hybridize with the hTR region may be considered as a method of inhibiting telomerase. GRN163L, which is known today as Imetelstat, is a 13-nucleotide sequence of lipidized N3’-p5’ thiophosphoramidate. The thio-phosphoramidate component makes this molecule soluble in water, stable against acids, and resistant to nucleases, and gives it the capability to form a stable RNA duplex. The 5’palmitoyl component of the GRN163L molecule causes its greater absorption by cells. The sequence of GRN163L (5’-palmitate-TAGGGTTAGACAA- NH2-3’) targets the 13-nucleotide region of the hTR, preventing it from forming an active complex with hTERT. Because telomerase inhibitors may require a period of treatment to produce telomeres short enough to trigger cancer cell death, telomerase inhibition therapy may be most effective when used in conjunction with conventional chemotherapies, radiation or other targeted therapeutics (39).
3.3. G-quadruplex Stabilizer
The 3’ overhang of human telomeres is guanine rich and can form characteristic secondary structures under physiologic ionic conditions, i.e. G-quadruplex structures. It has been hypothesized that these structures might be important for telomere function (40). G-quadruplex structure must first be unfolded before the telomerase can initiate extension of the telomere. Therefore, if telomeres could be stabilized using a G-quadruplex structure, the cells could be prevented from infinite proliferation characteristic of cancer. Small molecules that stabilize these structures and mimic their effect have been designed and found to inhibit telomerase activity. The three most commonly studied G-quadruplex stabilizing agents are telomestatin, BRACO-19 and RHPS4 (1).
3.4. Immunotherapy
The discovery of tumor-associated antigens that are universal to a broad range of tumor types would greatly enhance the efforts to target these antigens in strategies such as vaccination and in generation of effective anti-tumor Cytotoxic T Lymphocyte (CTL) responses. Telomerase is present in the majority of human tumors and is, therefore, a good candidate as a universal tumor-associated antigen (41). As an ideal tumor-associated antigen (TAA), hTERT not only possesses tumor specificity with little or no expression in differentiated healthy somatic cells, but also produces epitopes for both MHC class I and II pathways to induce “help” for CD8+ Cytotoxic T Lymphocyte (CTL) responses capable of attacking tumors. To boost the anti-telomerase immune response, 26 hTERT peptides have been exploited as epitope-mimicking structures (mimotopes) for anti-tumor immunity. Among these peptides, 23 are for MHC class I-mediated antigen presentation, capable of eliciting specific CTL responses, whereas three are for MHC class II mediated antigen presentation to induce CD4+ T cell responses. This suggests that hTERT antigenic peptides represent potential molecular targets for the treatment of cancer (42).
3.5. Telomerase-Directed Gene Therapy
One of the major goals in anticancer therapies is to target toxic agents to tumor cells specifically, to minimize the effects toward normal cells. The specific expression of telomerase hTERT in most types of tumors provides a good discrimination between cancer and normal cells. Deletion analysis of the hTERT promoter reveals a core promoter region located approximately 200 bp upstream of the transcription start site, and is sufficient to confer its specific expression in cancer cells. The property of hTERT promoter has been applied to restrict the expression of therapeutic genes in tumors. The utilized therapeutic genes include apoptosis-inducing, toxin-encoding, chemotherapeutic sensitizer and xenoantigen genes, or genes used in gene directed enzyme prodrug therapy (GDEPT). These hTERT promoter-driven therapeutic genes are introduced into tumor cells through liposome- or virus-mediated pathways. As expected from the expression pattern of telomerase, this type of approach selectively kills virtually all types of telomerase-positive cancer cells without affecting the viability of telomerase-negative cells in both cellular and animal xenograft models (43).
3.6. Problems With Telomerase Inhibitor
One of the problems with telomerase inhibitors is a phenomenon known as “phenotypic lag”. Telomerase inhibitors cause erosion of telomeric sequence over cell divisions until critically short telomeres signal senescence and apoptosis in cancer cells. Most solid tumors have a population-doubling time of several weeks, thereby months of treatment are required for the telomerase inhibitors to exhibit their effects. Hence, the more practical approach would be to develop reliable quantitative assays to measure distribution of the shortest telomere in a given cancer cell population so that oncologists can select cancer patients with more chances of responding to telomerase inhibitors (1).
4. Conclusions
From the time, when Elizabeth Blackburn identified telomerase and introduced the science of telomeres and telomerase, up to now, considerable progress has been made as many researchers have conducted research on its various aspects, such as the chemical and structural aspects, and on aging biology and cancer biology. In this article, the biological structure and the functions of telomeres and telomerase were reviewed and the presence of telomerase in cancer and somatic cells was discussed. Since telomerase is expressed in 90% of cancer cells, its suppression can be considered as a goal of cancer treatment. Various methods have been developed for telomerase suppression and obtained results indicate telomerase-based treatments may have the potential of forming the main part of cancer treatment methods.