1. Background
Recurrent spontaneous abortion (RSA) is one of the most common complications of pregnancy, particularly in the first trimester, which takes place in approximately 15% of clinically recognized pregnancies (1). Also, it is estimated to occur in almost 1% of all couples. Recurrent spontaneous abortion (RSA) is defined as the involuntary termination of two or more pregnancies before 20 weeks of gestation. Accumulating evidence suggests that recurrent spontaneous abortion is a complex multifactorial defect caused by various, genetic and non-genetic, factors which involves in cardiovascular disease and metabolic dysfunction. Etiologic factors such as genetics, anatomic, endocrine, placental anomalies, immunologic and hormonal problems, infection, lifestyle, smoking, and alcohol consumption are recognized as risk factors of RSA (2). Some previous studies revealed the relationship between genes involved in vasoconstriction, coagulation and angiogenesis with the risk of recurrent placental abruption. A normal pregnancy is related to adequate placental blood flow and fetal vasculature. Complicated cooperative interactions between different cell types and various growth factors in the processes of implantation, embryonic development and placentation are required for normal functioning vascular network development (3). Several gestational complications such as miscarriage, intrauterine fetal death, intrauterine growth restriction and preeclampsia may happen due to placental vasculature disorders (3).
MicroRNAs (miRNAs) are evolutionarily conserved small non-coding single-stranded RNA molecules with an average size of about 18 to 25 nucleotides in length. They are involved in post-transcriptional control of gene expression by mediating translational repression or mRNA degradation. The miRNAs play an important role in physiological and pathological processes. A growing number of microRNAs correlate with different human cancers, since they can function as tumor suppressors and oncogenes. Many major cellular functions and various regulatory pathways such as development, apoptosis, differentiation, cell proliferation, growth, and metabolism are regulated by miRNAs as signaling molecules (4, 5). In addition, human villous trophoblast secretes placenta-specific miRNAs in maternal circulation. It is also known that dysregulation of microRNA are involved in many diseases such as cancer, cardiovascular disease and complications of pregnancy such as preeclampsia. Therefore, it is likely that miRNAs are necessary to regulate genes involved in pregnancy (5). Previous studies show that placental abruption, the premature separation of placenta from the uterus and separation of the placenta before delivery may be related to variation in maternal cardiovascular and metabolic genes. Moore et al. suggested that MIR17HG gene (miR-17 ~ 92cluster) may contribute to placental abruption pathogenesis as well as cardiovascular diseases, through a similar mechanism (6). A polycistronic microRNA cluster termed miR-17-92 located in 13q31.3 in the human genome is among the best-studied microRNA clusters and encodes at least six mature miRNAs including miR-17, miR-20a, miR-18a, miR-19a, miR-19b and miR-92a. The miR-17 ~ 92 is highly conserved in all vertebrates. Ancient gene duplications generated two polycistronic miR-19-72 paralogs in mammals including miR-106b-25 and mir-106a-363 clusters (7, 8). MIR17HG is the host gene for the miR-17 ~ 92 cluster involved in the events essential for a healthy pregnancy such as cellular proliferation, invasion, differentiation, adhesion, apoptosis and angiogenesis (6). Therefore, MIR17HG is a putative candidate gene for association with placental abruption due to its functions. The transforming growth factor beta (TGF-beta) is a vital regulator of placental development and implantation. Since MIR17HG is involved in regulation of TGFB, it may contribute to placental abruption pathogenesis (9, 10).
2. Objectives
The current study aimed to investigate the relationship between rs6492538 polymorphism of MIR17HG gene and recurrent spontaneous abortion, which might be clinically useful as a marker to assess risks for recurrent pregnancy loss to provide good health care during pregnancy.
3. Materials and Methods
In this case control study, 100 females in the age group of 20 - 45 years old with at least two recurrent pregnancy losses (2-5, 7) referred to Tehran medical genetics laboratory, Tehran, Iran, were selected as the case group. The patients did not have any other known risk factors including environmental factors, anatomic anomalies, infections, immunologic factors, hormonal imbalances, chromosomal abnormalities and endocrine dysfunction. Also 100 healthy females aged 22 - 50 years with at least two successful deliveries (2-5, 7) without any miscarriages, endometriosis and infertility in their reproductive history were selected as the control group. Written consent was obtained from all subjects.
Five milliliters of blood samples of the subjects were collected in ethylenediamine-tetra-acetic acid (EDTA)-containing tubes and genomic DNA was extracted using salting out method; the extracted DNA was stored at −20°C until analysis. Briefly, polymerase chain reactions followed by RFLP were performed to detect the presence or absence of the rs6492538 polymorphism (Table 1).
| Polymorphism | Primers | PCR Product, bp |
|---|---|---|
| rs6492538 | 401 | |
| F: | 5'-GGCACTAATCTTTCTTGAACACTG-3' | |
| R: | 5'- ACCCAAGGTAAACAGAAGAGCAG-3' |
PCRs were performed with 80 ng of genomic DNA, 0.5 U/μL of Taq DNA polymerase (cinnaGen), 3μL of 10X PCR buffer, 1.5 mM MgCl2, 0.25 mM of all four dNTPs, and 5 - 7 pmol of each primer, in a total volume of 25μL.
After denaturation at 95°C for five minutes, the temperature was cycled 30 times (95°C for 50 seconds, 63°C for 50 seconds, and 72°C for 50 seconds), followed by extension at 72°C for 10 minutes to amplify the target DNA. Ten microliter of PCR product was digested overnight at 37°C with one unit of Alu1 restriction enzyme (New England biolabs) for rs6492538 polymorphism, respectively, in a final volume of 30 μL, according to the the manufacturer instruction.
The amplified fragment of 401bp for rs6492538(A/C) SNP was digested with Alu1 enzyme.
3.1. Statistical Analysis
The frequencies of alleles and genotypes for the studied SNPs in miR17HG gene were compared using the Chi-square test (SPSS 18.0). P values less than 0.05 were considered statistically significant.
4. Results
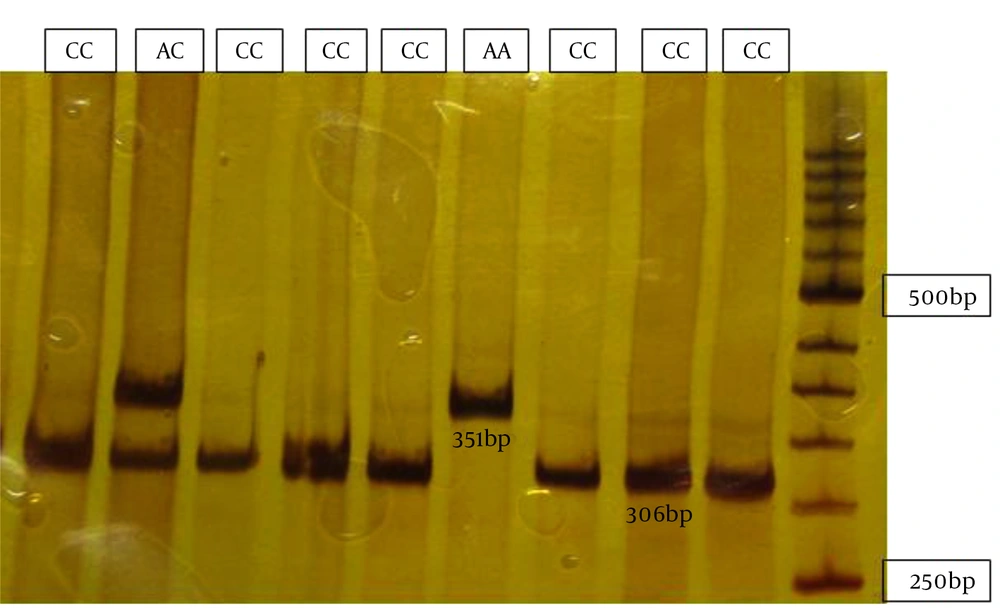
Individuals’ homozygous for the A allele showed two fragments of 351 and 50 base pairs. Individuals’ homozygous for the C allele showed three bands of 306, 50 and 45 bp. Those heterozygous for both the A and G alleles showed four bands of 351, 306, 50 and 45bp.
Digested fragments were separated by electrophoresis on 12% acrylamide gel stained with silver nitrate. In order to confirm the results, samples were randomly sequenced using 3130 ABI capillary electrophoresis (Figure 1).
The genotype and allelic frequencies obtained for the rs6492538 SNP are presented in Table 2.
aGenotype and allele frequencies between patients and control subjects were compared by the Pearson chi-squared test.
bP < 0.05 was considered statistically significant.
The frequencies of AA, CC, and AC genotypes in the cases were 4, 23, 73% and in controls were 14, 28, 58%, respectively (P = 0.007 and 0.08). The frequencies of A allele in the cases and controls were 15.5% and 28%, and the frequencies of C allele in the cases and controls were 84.5% and 72%, respectively (P = 0.002).
5. Discussion
Recurrent spontaneous abortion (RSA) can be physically, emotionally and economically hurt couples and societies. Therefore, identifying risk factors of recurrent miscarriage is of great importance and might provide exciting new therapeutic opportunities. Observations of high recurrence rates and familial aggregation of placental abruption in patients propose genetic predisposition to placental abruption. Genetic variations related to placental abruption such as polymorphisms in the genes involved in coagulation, the renin-angiotensin system, angiogenesis, inflammation, tissue remodeling, and homocysteine metabolism are identified in previous studies. The current study evaluated the relationship between the rs6492538 polymorphisms of MIR17HG and RSA. Previous studies investigated the effect of rs6492538 polymorphism of MIR17HG on premature separation of placenta from the uterus and cardiovascular diseases but not on placental abruption.
However the overlap in genetic mechanisms between placental abruption and cardiovascular diseases suggests that MIR17HG may provide exciting information as a candidate gene. Ota et al. firstly demonstrated that miR-17 ~ 92 cluster functions as an oncogene overexpressed in B-cell lymphoma cell lines and diffuses large B-cell lymphoma patients with 13q31-q32 amplifications (11). Studies on miR-17 ~ 92 cluster functions are continued. Dews et al. showed the role of miR-92a overexpression on inhibition of tube formation, cell migration, and adhesion of endothelial cells; while it had no effect on cell survival and proliferation. Accordingly, inhibition of miR‐92a with antagomirs induces angiogenesis and improves recovery of blood flow in hind limb or myocardial ischemia resulted in tissue viability in the region (12). Ventura et al. studied the mir-17 ~ 92 and its paralogous clusters, mir106a-363 and mir106b-25, and reported that deletion of mir-17 ~ 92 gives rise to defective embryonic development of fetal heart, lung and B-cells and postnatal death (13). Smith et al. revealed a 25% and 56% increase in risk of ischemic heart disease (IHD) among the parents of females with two previous losses and parents of females with three or more losses respectively (14). Hence, common pathophysiological pathways and genetic predispositions may contribute in recurrent miscarriage and IHD. Fu et al. indicated that MIR17HG gene (miR-17 ~ 92 cluster) contributes to the pathogenesis of preeclampsia as a potentially dangerous pregnancy complication including recurrent pregnancy loss (15). Reports suggest that since MIR17HG is involved in regulation of TGF-beta, which regulates the placental development and implantation, may be involved in placental abruption pathogenesis (9). Ventura et al. reported that miRNA-17and miRNA-19b, members of the microRNA-17-92 cluster, are expressed in early stages of pregnancy. Interestingly, down-regulation of microRNA-17 and -19b are observed in villous samples from early miscarriages (16). It is suggested that these microRNAs might contribute to placental invasion. Dysregulation of miRNA-17 and miRNA-19b might be associated with defective placentation. Moore et al. identified placental abruption as a complex multifactorial pathogenesis which is more likely to share common pathologic mechanisms responsible for cardiovascular and cardio-metabolic diseases. Based on similarities in the etiology of both conditions, they presented some genes involved in cardiovascular disease that may be related to placental abruption (8). Moore et al. reported the association between two SNPs of MIR17HG, including rs4773624 and rs6492538, and placental abruption (8). Based on available reports MIR17HG is essential for angiogenesis, cell survival, development, apoptosis and other pivotal processes; therefore, it could be considered as a candidate gene to investigate recurrent abortion.
The current study investigated the relationship between the rs6492538 polymorphism of MIR17HG and RSA, which might be clinically useful as a marker to assess risks for recurrent pregnancy loss. Results of the present study showed that rs6492538 and CC genotypes are contributing factors in PRL; in line with those of the study by Moore et al. and there was no other studies in this filed to compare (8).
Considering the results of the study, the hypothesis of the relevance of this factor with the RPL is still unanswered. Therefore, to investigate this factor and its detailed association with RPL, further cohort studies with larger sample sizes are suggested. It is recommended that subjects be selected from the same race and region. Risk factors including hypertension, weight, cardiovascular disease history, occupation and continue working during pregnancy, caffeine consumption and alcohol use should be considered.