1. Background
Targeting DNA by small drugs modifies and/or inhibits the functions of DNA in cells. The two interactions suggested for drug binding to DNA are irreversible covalent, and reversible and non-covalent bonds. The covalent form induces cell death through inhibition of DNA’s functions. Non-covalent drug-DNA interaction has been classified into three types including intercalation, groove and external bindings. These interactions can change DNA conformation and torsional tension, and also dissociate protein-DNA interactions lightly and may break DNA strands (1, 2). There are various anticancer agents derived from herbs that interact with DNA such as mitoxantrone (3), curcumin (4), quercetin (5) and saffron metabolites (6, 7).
Rosmarinus officinalis belongs to the Lamiaceae family, popularly known as rosemary (8). The main compounds of rosemary are rosmarinic acid, carnosic acid, carnosol, rosmanol, flavonoids and other phenolic compounds (9). This herb has many medicinal properties including antibacterial, antioxidant (8) and anticancer (10, 11). Rosemary has been shown to inhibit the proliferation of several human cancer cells such as breast, leukemia, prostate, lung and liver (10).
2. Objectives
Altogether the information about molecular mechanisms of anticancer effect of rosemary is very limited. Thus, we designed this study to investigate the interaction of rosemary with high molecular weight DNA.
3. Materials and Methods
Rosemary used in present study grows in Birjand, south Khorasan with herbarium code of 28. To prepare rosemary aqueous extract, the flowers of the plant were washed and dried at 50°C. Five grams of plant powder was blended with 100 mL of boiling water and brewed for 30 minutes. The solution of the plant was centrifuged at 7000 g for 30 minutes. After filtering of the supernatant by Whatman No. 1, the obtained sample was lyophilized by freezing at -80°C for two hours. High molecular weight DNA was purified from calf thymus, as previously illustrated. The ctDNA (calf thymus DNA) concentrations were determined by an extinction coefficient of 6600 M-1cm-1 at 260 nm and expressed in terms of base molarity (7).
Titration of ctDNA (0.03 mg/mL) and buffer was carried out at various concentrations of rosemary extract (0 - 16 mg/mL) and EtBr (ethidium bromide) source (0.008 mg/mL) for one minute in each addition. All the spectrophotometric measurements were accomplished by an Eppendorf spectrophotometer (USA).
Fluorometric measurements were performed using a Shimadzu Model RF-5301 spectrofluorometer. The spectra record was at fast scanning speed. The excitation and emission wavelengths of rosemary extract was determined at 410 and 494 nm, respectively.
Circular dichroism (CD) measurements were done on a JASCO model J-715 CD recorder at 25°C. Results are reported as molar ellipticity, [θ] × 10-3 (degree. Cm2/dmol), based on the average weight of nucleotide (AWN), which was equal to 330 for DNA. The molar ellipticity was characterized as:
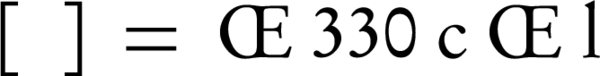
where ‘c’ is the concentration of DNA in mg/mL, ‘l’ is the light path length in centimeters, and ‘θ’ is the record ellipticity in degrees at a wavelength of λ. All experimental interactions were performed in 0.05 M tris buffer 7.4, at 25°C.
4. Results
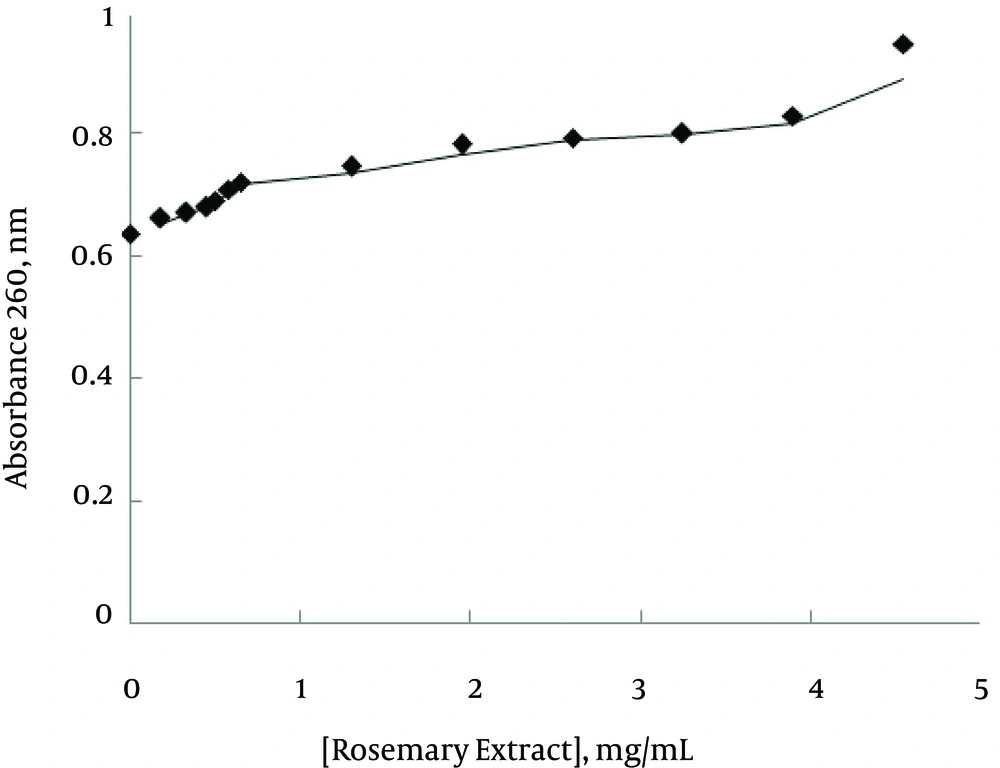
The spectrophotometric titration of ctDNA with rosemary extracts was used in order to determine the interaction between ctDNA and rosemary extract. The nonlinear increment in the absorbance of ctDNA at 260 nm was observed with increasing rosemary extract concentration (Figure 1).
The difference in the fluorescence intensity of the rosemary extract in the absence and presence of ctDNA were shown as fluorescence emission (Figure 2). The fluorescence emission of rosemary extract indicates its interaction with ctDNA.
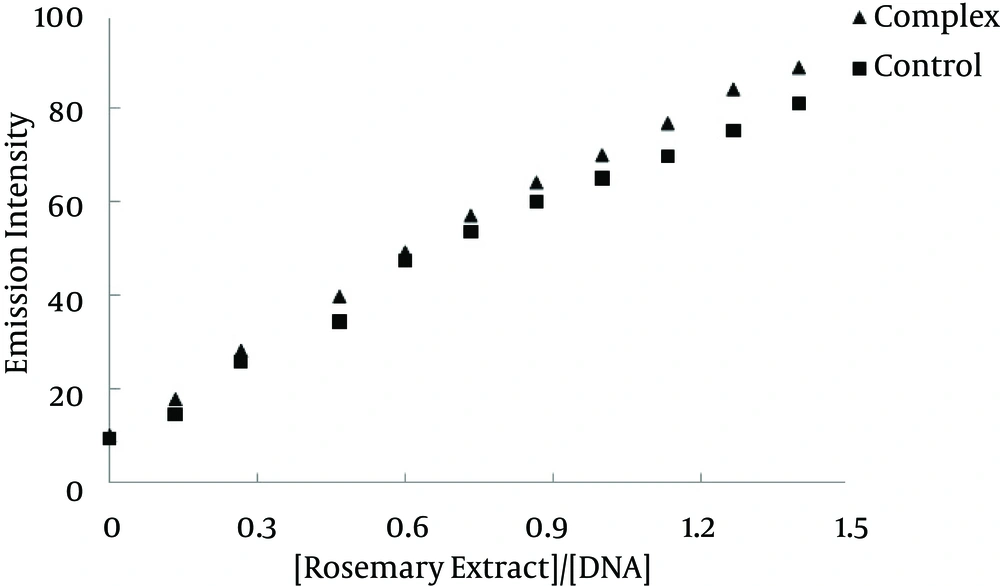
Moreover, the fluorescence quenching of EtBr bound to ctDNA by rosemary is illustrated in Figure 3A. This effect of rosemary extract is in agreement with the linear Stern-Volmer plot (Figure 3B). The Stern-Volmer constant (K) value for rosemary extract is 0.39 (mg/mL)-1. In the plot of F0/F versus [rosemary extract]/[DNA], K is achieved by the ratio of slope to intercept (12).
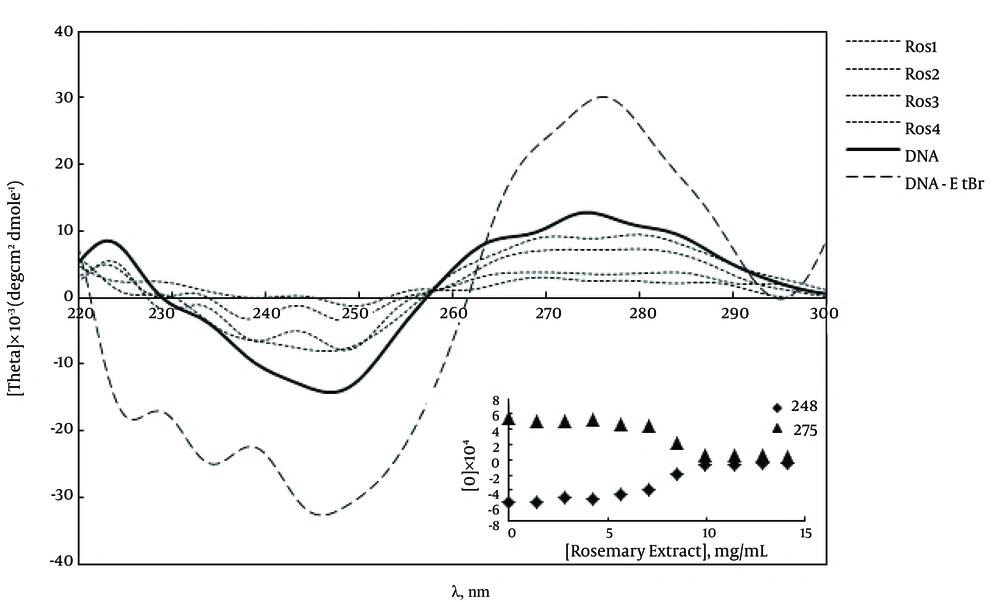
The CD spectra of ctDNA in the presence of rosemary extract and EtBr (as a control) were plotted. As indicated in Figure 4, the CD spectrum of DNA in the presence of EtBr shows an increase in the positive band at 275 nm, and the negative band at 248 nm, which are the characteristic peaks of an intercalating agent. However, in the presence of rosemary extract, these typical peaks are not observed and the CD spectra of DNA are significantly perturbed.
The CD spectra at various concentrations of rosemary extract are shown in Figure 4. In comparison with the characteristic features of the B-form DNA, which are seen in each Representation , some changes are observed in the CD plots of DNA in the presence of different concentration of these ligands. Changes include decrement in the peak at 275 nm and reduction in the negative value at 248 nm. At higher rosemary extract concentrations, the precipitate was formed and the spectra were distorted completely.
5. Discussion
Currently, many anticancer drugs can interact with the double stranded DNA. These drugs exhibit cytotoxic activity on tumor cells via preventing DNA replication and transcription or inhibiting gene expression (13). Rosemary extract has been shown to have significant anti-proliferation effects on various human cancer cells (10), yet its molecular mechanism is unclear. In this context, we studied the interaction of rosemary extract with ctDNA as one of the possible mechanisms for its anticancer property.
In the present study, UV-Vis spectrophotometries were initially used to investigate the formation of rosemary extract and ctDNA at 260 nm absorption band. It is well known that intercalation of compounds into DNA leads to hypochromism and bathochromic shift (14), which is different with the observed hyperchromism of rosemary-ctDNA interaction. Besides, this hyperchromic effect may reflect the conformational changes or distortion of DNA structure due to interaction with rosemary (14).
Fluorescence and CD spectra of DNA in the presence of rosemary revealed the proposed mechanism of interaction. The fluorescence intensity of rosemary extract emitted in the presence of ctDNA, which indicated the complete rosemary-ctDNA interaction. On the other hand, fluorescence intensity of EtBr emitted in the presence of DNA was due to its strong intercalation with adjacent DNA base pairs. It was previously shown that the increased fluorescence could be quenched by the addition of a second molecule (12). Also rosemary extract quenched the emission spectra of EtBr bound to ctDNA. Such behavior has been previously reported for two synthetic water-soluble porphyrins that bind to DNA by out-side self-stacking along the DNA helix. The extent of fluorescence quenching of EtBr bound to DNA was applied to determine the extent of binding between the second molecule and ctDNA (12). The Stern-Volmer Equation was was as follows,
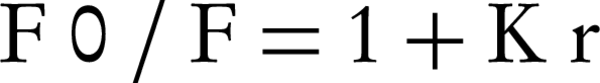
Where F0 and F are the fluorescence intensities in the absence and presence of complex, respectively, K is a linear Stern-Volmer quenching constant and r is the ratio of total concentration of complex to that of DNA (12); the Stern-Volmer constant value for rosemary extract was 0.39 (mg/mL)-1. Since, EtBr intercalates DNA through interaction with the minor groove; the displacement of EtBr by the titration of a second molecule is suggestive of an intercalative or minor groove binding (15).
These data are the affirmative reason for the results obtained by CD. Circular dichroism spectroscopy measures the difference in the absorption of left and right circularly polarized light, so this technique is used to study of conformational changes of DNA due to ligands addition (13). Rosemary extract causes changes in the CD spectra of ctDNA including decrease in the positive peak at 275 nm and negative peak at 248 nm. These changes indicated some conformational changes in ctDNA structure due to the B to C-form transition (7). Also, at higher concentrations of rosemary extract, positive and negative peaks come near to the zero point. It indicated the irregular changes base-base interaction due to the un-stacking of DNA base. Such behavior has previously been reported for ingredients of saffron (6, 7).
As mentioned in the introduction, the rosemary extract contains different components such as carnosic acid, carnosol and rosmanol. Thus, each of these components may interact with DNA via various mechanisms such as intercalation, groove binding and external binding. Our results of spectroscopic studies show that rosemary extracts interact with ctDNA. Our results indicated that the likely major mechanism for ctDNA-rosemary extract interaction is minor groove binding; because the extract is a mixture of ingredients, we cannot talk confidently about it. Therefore, we recommend further investigations on the interaction of all components of rosemary extract with DNA to clarify the exact mechanism.
![A, emission spectra of EtBr bound to DNA in the presence of the rosemary extract; B, fluorescence quenching curve of EtBr bound to DNA by the rosemary extract. [EtBr] = 0.008 mg/mL, [DNA] = 0.03 mg/mL, [rosemary extract] = 0 - 0.042 mg/mL. A, emission spectra of EtBr bound to DNA in the presence of the rosemary extract; B, fluorescence quenching curve of EtBr bound to DNA by the rosemary extract. [EtBr] = 0.008 mg/mL, [DNA] = 0.03 mg/mL, [rosemary extract] = 0 - 0.042 mg/mL.](https://services.brieflands.com/cdn/serve/3170b/2d9457dd6e0b0ec5dc1bdfe9ac282c7943fbda06/gct-In_Press-InPress-35638-i003-preview.webp)