1. Context
A new era in biology began in 1970 with the development of recombinant DNA technology. This breakthrough allowed biological researchers to modify DNA molecules for the first time, opening up new therapeutic possibilities. Today, targeted genome modification techniques offer a powerful tool for treating genetic illnesses using living cells. One of the most recent methods for genome editing is the clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 technique, which draws inspiration from the bacterial immune system's gene-editing capabilities. This method is renowned for being rapid, simple, and highly effective.
Recent advancements in CRISPR technology, such as the integration of antisense RNA and metal/metal oxide nanoparticles, are expanding its applications across various fields (1). These innovations are enhancing not only the speed and accuracy of diagnostic methods, such as those used for detecting SARS-CoV-2, but also CRISPR-Cas9’s potential in targeting pathogenic bacteria. These developments highlight CRISPR's increasing role in revolutionizing medical diagnostics and therapeutic strategies, as well as its applications in medicine and stem cell engineering. By utilizing these new methods, researchers are advancing the precision and effectiveness of treatments for genetic diseases and bacterial infections (2).
This article explores the theoretical foundation of CRISPR technology and its applications in treating genetic diseases. The primary aim of this technique is to remove, add, or alter specific genes within a cell. Since the initial discovery of the CRISPR system's ability to edit genomes precisely, it has been recognized as a simple yet powerful tool (1, 3, 4). The discovery of CRISPR-Cas systems and their use in genome engineering has fundamentally transformed research in life sciences (4). The CRISPR-Cas system enables precise, RNA-dependent genome editing, widely used in fields like biology, medicine, and gene therapy. However, the efficiency of gene editing can vary depending on the guide RNA used, and there is the risk of both on-target and off-target cleavages, leading to unintended genetic changes. To address these challenges, several command-line and web-based tools have been developed to help scientists select optimal CRISPR target sites for different systems, such as CRISPR-Cas9 and prime editing tools.
It is crucial to assess the mutation rates and patterns that arise following CRISPR treatment in cells or organisms (5). Sanger sequencing and high-throughput sequencing are among the most accurate methods for analyzing genome editing outcomes. Numerous tools exist for analyzing data from next-generation sequencing (NGS) (6). This article discusses the latest target design and analysis tools for genome editing, emphasizing the significance of CRISPR in advancing medical research and stem cell applications, and outlining its potential for future innovations.
2. Evidence Acquisition
2.1. Date and Mode of Searching the Articles
This research examined papers from 2007 to 2024 that were indexed in ISI, SID, PubMed, and PubMed Central. The review was limited to original English articles, with a special focus on summarizing the investigated medical applications, stem cell engineering, the latest target design or analytic programs for genome editing, and advances as well as obstacles.
2.2. Searched Keywords
Genetic Therapy AND Cancer AND RNA-guided Engineered Nucleases AND Gene Editing, Regenerative Medicine AND Embryonic Stem Cells AND Zinc Finger Nucleases.
3. Results
Several cross-checks and the removal of cases that did not meet the criteria were involved in selecting the most relevant articles. Important insights were gathered from the search results obtained from various databases. Of the 36 articles reviewed, 29 related articles were selected. To ensure accuracy and reliability, a thorough evaluation process was applied to the selected articles. Based on research findings, the CRISPR technique can be used to correct specific genetic defects in patients during gene therapy, providing a treatment option for diseases that were previously incurable through traditional methods.
3.1. Stem Cell
Stem cell therapy has developed into a highly promising and sophisticated area of scientific study in recent years. The human body contains unspecialized stem cells. The development of clustered regularly interspaced short palindromic repeats (CRISPR) and induced pluripotent stem cells (iPSCs) represents a significant breakthrough in drug development and regenerative medicine (7). These autologous stem cells can be controlled through biological, chemical, or genetic methods to fix gene abnormalities or activate preprogrammed cues to direct cell reprogramming or differentiation, eliminating the need for immunosuppression. Additionally, the capacity of autologous stem cells to multiply, differentiate, and produce cytokines and growth factors to promote tissue regeneration and alter the local microenvironment makes them a key cell source in individualized regenerative medicine. Numerous sources of stem cells, including the embryo, bone marrow, adipose tissue, blood, and others, can be used to isolate various types of stem cells. Pluripotent stem cells (PSCs), such as ESCs and iPSCs, can differentiate into nearly all types of cells, including those that are challenging to isolate and grow in quantity (such as neurons) (7). ASCs and BMSCs are commonly utilized in promoting the growth of bone and cartilage because they have the ability to develop into adipocytes, chondrocytes, or osteocytes. HSCs are excellent for treating blood diseases because they can differentiate into every type of hematopoietic cell (8).
3.2. Genome Editing
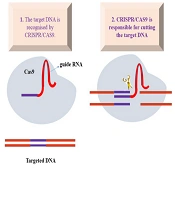
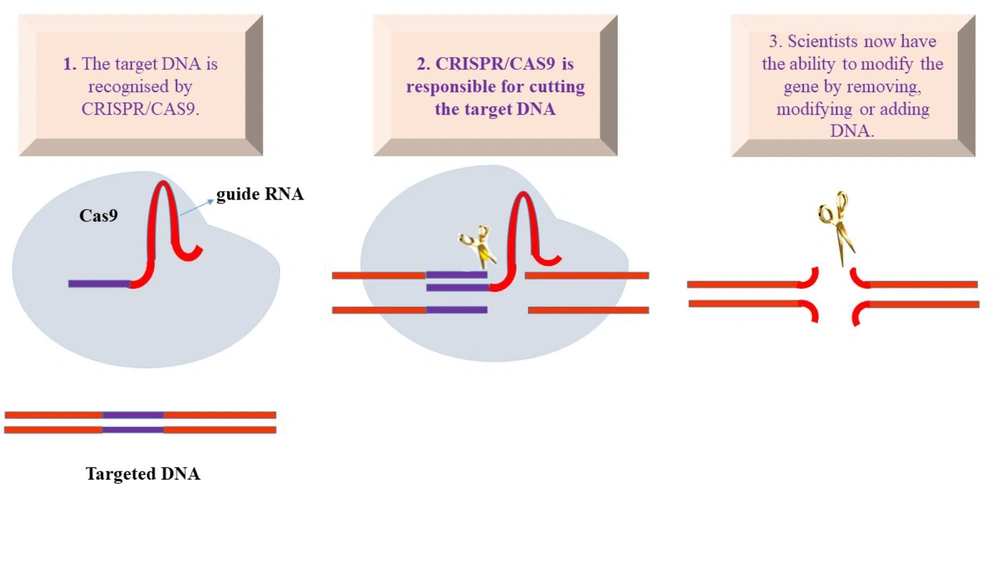
Genome engineering refers to the deliberate insertion of genetic components to identify genes that can be targeted, either to disrupt their sequence leading to a decrease in the activity of the gene or to modify the protein sequence produced by the transcribed gene. ZFNs and TALENs have been replaced by CRISPR/Cas systems as an effective and theoretically simpler option for producing DSB-mediated gene changes (9). The most well-known RNA-guided engineered nucleases (RGENs) are the bacterial and archaeal CRISPR and CRISPR-associated systems. Programmable nucleases also include the first truly targetable zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and the popular TALENs (10). Additionally, Cas9 nickase, which contains a single nuclease domain that has been deactivated, was developed to cause specific single-strand DNA nicks. Cas12a, produced by many species, has been used to modify the human genome (11). Figure 1 illustrates the use of the two-part CRISPR system for gene editing, where an enzyme called Cas9 guides DNA by cutting it according to the guide RNA.
3.3. Clustered Regularly Interspaced Short Palindromic Repeat Technology and Its Mechanism of Action
Since its discovery, CRISPR systems have been primarily used for genome editing, stemming from the finding that Cas proteins can make precise DNA cuts. The type II Cas9 protein from S. pyogenes was the first protein demonstrated to achieve this in a straightforward manner (12). Efforts are underway to explore whether other CRISPR Cas9 protein types can be employed for gene editing, particularly with the emergence of Cas9 orthologs and SpyCas9 variants. Although type I CRISPR systems were the first to be thoroughly studied, their requirement for several proteins (at different stoichiometries) made their use in eukaryotic systems less appealing. The implementation of type I systems for editing in human cells is relatively recent, although type I systems are widely used in microbial engineering (13). Another popular CRISPR system is Cas12a (formerly known as Cpf1), a component of type V systems, which offers several advantages over Cas9 proteins. Three Cas12a proteins have been described, including those from Francisellanovicida (FnCas12a), Acidaminococcus sp. (AsCas12a), and Lachnospiraceae bacterium (LbCas12a). These proteins use T-rich PAM sequences such as TTN and TTTN, distinguishing them from Cas9 proteins that utilize G-rich PAM sequences. Unlike Cas9 proteins, which require tracrRNA, Cas12a proteins can process crRNA from pre-crRNA independently (14). This feature has been utilized to enhance simultaneous genome editing at multiple loci by expressing several Cas12a-compatible crRNAs as a single pre-crRNA transcript, instead of multiple expression modules. However, the main advantage of Cas12a is its cleavage mechanism, producing staggered endpoints outside the critical seed region, unlike the blunt ends produced by most Cas9 proteins (15). The research discussed involves conducting extensive CRISPR functional genomics studies that simultaneously alter the function of multiple genes to systematically identify genes that influence biological processes related to disease characteristics. Epigenetic editing, which can be highly specific and easily adjusted, allows for the exploration of functional genomics across the entire genome with high accuracy, helping to identify true positives and minimize false hits (16).
The initial loss-of-function at the genome scale using dCas9 KRAB was the focus of CRISPR interference (CRISPRi) functional genomics investigations aimed at inhibiting the expression of protein-coding genes. These studies yielded expected results, including the identification of genes encoding ribosomal components, spliceosomes, and DNA replication machinery proteins, which are often essential for survival or cell proliferation (17). The first genome-scale gain-of-function study using CRISPRa, in a functional genomics context, revealed key findings in a screen for cell survival and proliferation, such as the identification of tumor suppressor genes, factors that regulate gene expression and influence cell status, and mechanisms that control the cell cycle apparatus. Tumor suppressor genes must serve as predicted positive control genes for CRISPR-based screens conducted in proliferating cells. During CRISPR-based functional genomic screening, increased expression of tumor suppressor genes leads to cell death or inhibition of cell cycle progression, resulting in the deletion of sgRNAs targeting tumor suppressor genes (18). Fusion genes are identified in solid tumor types, and due to their tumor-specific expression, they hold significant potential as therapeutic targets (19). Despite the importance of fusion genes in cancer, it has been challenging to model chromosomal rearrangements. The echinoderm microtubule-associated protein-like 4 (EML4) and anaplastic lymphoma receptor tyrosine kinase (ALK) genes were connected by an intrachromosomal inversion, which was the first fusion gene identified in lung adenocarcinoma (20). Two groups developed a CRISPR/Cas9-based technique to induce specific chromosomal rearrangements and generate an Eml4-Alk gene fusion in vivo. This was achieved by using Cas9 and two sgRNAs to target the intronic regions of Eml4 and Alk, enabling a concomitant DSB. The resulting tumors responded to crizotinib treatment, an approved ALK kinase inhibitor, making those models clinically relevant (21). Another team explored the use of the same method to induce various chromosomal rearrangements in HEK293 cells, leading to the creation of novel fusion genes that play a key role in lung cancer. They demonstrated the generation of intrachromosomal inversions, both paracentric and pericentric, as well as the unexpected promotion of interchromosomal translocations. However, further research is required to determine how effectively such techniques can induce translocations in vivo (22). Clustered regularly interspaced short palindromic repeat /Cas9 components have been delivered in vivo using recombinant adeno-associated viruses (AAV) and recombinant adenoviral viruses (AdV). Pooled CRISPR screening techniques enable large-scale, concurrent interrogation of many genotype-phenotype relationships within a single cell population. Briefly, titrated lentiviruses are used to introduce CRISPR/Cas9 libraries into a pool of cells. These cells can then be enriched for a specific phenotype, and NGS is used to identify the genes involved in that phenotype (23).
The CRISPR reagents are delivered by lentiviruses, which integrate stably into the genome, allowing the phenotype to be linked to a specific disruption within a cell. As a result, sgRNAs or gene knockouts that influence the desired phenotype can be identified through sequencing of sgRNAs in downstream cells within a population that has been specifically targeted (24). Modified CRISPR systems can precisely and reliably modify the transcriptome and epigenome. These technologies have significantly improved our ability to conduct experimental searches for genes that influence disease phenotypes (25).
3.4. Use of Clustered Regularly Interspaced Short Palindromic Repeat -Edited Human Being Stem Cells
While some mammalian cell lines can be edited with CRISPR, most primary cells are generally resistant to the technique. For example, human PSCs are more challenging to engineer using the CRISPR method compared to mouse ESCs or tumor cell lines (26). Genome editing of human PSCs using CRISPR is hindered by DSBs, which activate the TP53 gene, leading to cell cycle arrest, cell death, and other processes. The production of engineered human PSCs may be facilitated by TP53 suppression, which could also improve engineering efficiency. However, TP53 inhibition may simultaneously increase off-target mutations and raise cancer risk (27).
CRISPRa and CRISPRi enable tissue regeneration and stem cell engineering. In regenerative medicine, achieving robust expression of components required for cell differentiation using CRISPRa to reach optimal gene activation levels for cell programming is challenging. CRISPR interference, when used in conjunction with CRISPRa, inhibits differentiation into a competing lineage (such as adipogenic), while CRISPRa promotes differentiation into the desired lineage (28).
Finally, it has been found that modern CRISPR/Cas13 systems (such as Cas13a and Cas13d) contain RNase domains that facilitate target RNA cleavage in human cells. In addition, CRISPRi systems based on dCas9 and dCas12a restrict gene transcription at the DNA level (29, 30). Similar to RNA interference but with greater specificity, this approach may be used to control gene expression during the post-transcriptional stage. Further research is needed to determine whether this method is suitable for regenerative medicine and stem cell engineering (31, 32).
As previously noted, the limited availability of patient-derived tissues poses a constraint on biological systems, such as organoids derived from adult tissues. To develop useful cellular models, an alternative method is to differentiate PSCs into functional cell types that can partially replicate a target tissue. The use of human ES cells is difficult and strictly regulated, although they hold great promise (32, 33). When Takahashi et al. first demonstrated that adult differentiated cells can be reprogrammed into iPS cells, they opened up vast possibilities for use in areas such as disease modeling and personalized therapy. They showed that adult differentiated cells could gain pluripotency by overexpressing four transcription factors in human skin cells (34). This discovery was immediately recognized as a major medical breakthrough with the potential to revolutionize cell therapy, and it has already drastically changed drug discovery and preclinical research (35).
Multiple research groups have established and refined protocols to induce the differentiation of iPSCs into various types of mature cells. Human iPS cells are now used to model development or disease in several organs (36-38). Additionally, scientists have employed gene editing in human iPSCs to enhance or create new cellular disease models. More importantly, gene editing enables the establishment of isogenic control cell lines for iPS cell lines, whether from patients or healthy donors, or for mutant iPS cell lines. Induced pluripotent stem cells derived from patients with diseases caused by known genetic abnormalities serve as highly useful disease models, but scientists also require appropriate "control" iPS cells for comparison (37).
4. Discussion
A 2013 publication demonstrated CRISPR/Cas9's ability to accurately edit mammalian genomes, achieving precise genetic modifications in animal cells through CRISPR editing techniques (39). Since then, there has been an increase in the variety and efficiency of genome editing methods, which have been widely adopted by pharmaceutical industries and academia (40). Clustered regularly interspaced short palindromic repeat /Cas is a component of the bacterial immune system that functions to identify and eliminate foreign nucleic acid sequences (41). The frequently used CRISPR laboratory system utilizes the Cas9 nuclease derived from Streptococcus pyogenes (SpCas9). Clustered regularly interspaced short palindromic repeat /Cas9 technology is straightforward to use in laboratories, unlike TALENs and Zinc Finger technologies (29). Alongside the development of SpyCas9 and Cas9, there has been a surge in research to determine whether other forms of CRISPR Cas9 proteins could be used for gene editing. Although type I systems were the first CRISPR systems to be extensively studied (13), the requirement for multiple proteins reduced the appeal of employing this technology in eukaryotic systems. Type I systems have only recently been utilized for editing in human cells, although they are extensively used for precision engineering in microorganisms. Editing via the type I system is more difficult since it depends on a highly active Cas3 helicase/nuclease (30). Altering two specific amino acids in the RuvC and HNH domains has been found to disable the enzyme's catalytic function while maintaining its ability to bind DNA (42). This discovery enables the creation of a DNA-binding protein guided by sequence-specific sgRNA, providing a new opportunity for utilizing the bacterial immune system. By attaching a specific protein domain to a Cas9 enzyme that lacks catalytic activity, it becomes possible to target the activity of a protein to specific areas of the genome (33). This method was developed by linking dCas9 to repressive transcriptional domains, resulting in a potent 25 - 100-fold reduction in the expression of the targeted gene with no direct off-target effects. This method is also known as CRISPRi (17).
Commercial suppliers offer advanced tools and solutions for every phase of the CRISPR genome editing workflow. However, due to its size, Cas9 transport to cells poses a bottleneck for effective gene editing, particularly in vivo. The introduction of Rosa26 Cas9 mice, which express Cas9 constitutively, improved editing efficiency by reducing the components needed to just a small sgRNA (43). Additionally, Rosa26 loxP-Stop-loxP Cas9 knock-in mice and Cre mice, which express Cas9 constitutively upon activation, have also been developed (44). However, uncontrolled Cas9 expression may lead to off-target effects and inflammatory responses, limiting the usefulness of constitutive Cas9 systems. Although Wu et al. (45) emphasize that constitutive Cas9 expression following Cre induction in loxP-Stop-loxP Cas9 mice can have unforeseen cellular effects, Platt et al. argue that Cas9 is not harmful in the CrsCas9 mouse (38).
A primary goal of oncology research is to functionally identify the significance of genetic diversity in cancer, aiding researchers in developing more effective treatment strategies and guiding future therapies. In recent years, cancer medications have faced the highest failure rates in clinical development. Identifying the genetic underpinnings of tumor vulnerabilities may lead to the discovery of novel therapeutic targets, backed by genetic data and clear molecular mechanisms, thereby increasing the likelihood of success in clinical development and improving cancer treatment prospects. Genome-wide CRISPR/Cas9 screening is used to identify essential genes (46). Therefore, targeting these genes or proteins with specific drugs would reduce tumor viability. Extensive CRISPR screening is conducted using numerous cancer cell lines from various disease types (47). The identified genes are either crucial for basic cellular functions or play a key role in particular cancer cell types or environments (48).
The rapid technological progress of CRISPR makes it difficult to keep up with all developments. It is challenging to predict precisely what the future holds for CRISPR-based in vivo models. However, CRISPR/Cas9 tools are advancing the development of precise disease models, leading to improved treatment options for patients.
Although CRISPR technology is highly versatile, it comes with several limitations that must be considered in research and therapeutic applications. One of the main challenges is CRISPR's potential for off-target effects, where unintended genome cuts occur, posing significant risks such as disrupting essential genes or activating oncogenes. Delivering CRISPR components to target cells remains difficult, with current methods like viral vectors and lipid nanoparticles facing issues related to efficiency, safety, and immune responses. Additionally, CRISPR's editing efficiency is not always 100%, leading to a mixture of edited and unedited cells, which complicates therapeutic applications. Editing certain cell types, such as primary or stem cells, also presents challenges. Ongoing research aims to improve CRISPR's precision, delivery methods, and address ethical concerns (2). Despite these limitations, CRISPR holds immense potential for precise gene editing, facilitating breakthroughs in treating genetic disorders and driving innovation. Continued advancements aim to overcome the current challenges and enhance both accuracy and safety.
4.1. Conclusions
Since its discovery in 2012, the CRISPR system has become a cornerstone technique in laboratories worldwide, revolutionizing fields such as medicine, stem cell engineering, and drug discovery. By providing a precise, efficient, and versatile tool for genome editing, CRISPR enables the targeting and modification of specific DNA sequences, offering immense potential for treating genetic disorders, developing personalized medicine, and advancing regenerative therapies. In stem cell research, CRISPR-based genetic screening plays a crucial role by allowing scientists to explore a wide range of biological processes and uncover the genetic foundations behind them, thereby identifying disease causes and potential therapeutic targets. New companies are leveraging CRISPR to develop therapeutics, create diagnostics, and study diseases using cellular and animal models. As research advances and applications expand, CRISPR brings new hope for curing previously untreatable diseases and driving innovations in gene therapy and tissue engineering. However, ethical considerations and thorough safety evaluations are still necessary to fully explore its potential.