1. Background
Schizophrenia (SCZ) is a disorder with a prevalence rate of 1.1% in the population over the age of 18. Men tend to develop SZC symptoms earlier than women but frequency of occurrence in both sexes is the same (1, 2). Combined genetic predisposition and environmental exposure play a significant role in outbreak of this disorder (3). Genome and environmental interaction may provoke occurrence of neurobehavioral disease through DNA methylation in the regulatory region at the promoter sites (CPG islands) (4). Apolipoprotein E (APOE) is a Polymorphic Plasma Protein with three common isoforms E2, E3, and E4 of the APOE gene, which is situated on chromosome 19q13.21. There is a functional difference between 5' and 3' CpG islands of APOE, because APOE CpG island is methylated in all tissues except sperm but this gene is expressed in many tissues in terms of location of CPG island within the last exon of the gene (5). APOE protein as a lipid transporter is the main regulator of blood lipid in humans and plays an important pleiotropic function in the development of the central nervous system and synaptic plasticity (6). It is also involved in various pathways that are unrelated to lipid transport including suppression of T cell proliferation, regulation of macrophage function, facilitating of lipid antigen presentation by CD1 molecules to natural killer cells, and modulation of inflammation and oxidation (7, 8). Therefore APOE could be one of the candidate genes for long term impaired inflammatory reactions in schizophrenia patients.
Saiz PA et al. (2002) results showed that variation in the APOE gene expression indicate insignificant relationship with the development of schizophrenia in a Spanish population (9). By comparison, allelic variations of APOE gene seem to be momentous in pathogenesis of schizophrenic disorders in the Korean population (10). Schurhoff F et al. (2003) observed a possible association between male schizophrenic patients and the APOE epsilon 2, epsilon 3 genotype in the French population (11).
Carnitine palmitoyltransferase in mammals shows three isoforms; a liver isoform CPT1-A, a heart/skeletal muscle isoform CPT1-B and CPT1-C which exists in the endoplasmic reticulum of hippocampus neurons. These are the products of three different genes which are located at 11q13 (CT1-A), 22q13.3 (CPT1-B) and 19q13 (CPT1-C) chromosomes (12, 13), respectively. The CPT1-A gene provides instructions for making a mitochondrial enzyme called carnitine palmitoyltransferase 1A, which is found in tissues involved in glucose sensing, such as liver, pancreatic β-cells and hypothalamic neurons (14). Carnitine palmitoyltransferase (Cpt) mediates fatty acid translocation in β-oxidation process across the mitochondrial membrane which impairs it in its task and has simultaneous effects on fatty acid and glucose metabolism that can lead to a number of features of the chronic SCZ, such as hypoglycemia or hypoketosis which cause brain damage due to reduced energy production (15-17). Additionally, increased β-oxidation and decreased fatty acid/lipid synthesis may be associated with down regulation of myelination-and oligodendrocyte-related genes in SCZ (18).
2. Objectives
The goal of the present study was to investigate the effects of the promoter “hypermethylation” of APOE and CPT1-A and their expression profiles in the development of SCZ.
3. Patients and Methods
3.1. Study Subjects
This case–control study was conducted on 80 newly diagnosed and untreated SCZ individuals with mean age of 47.53 ± 10.801 and average onset age of 8.729 ± 20.79, who were admitted to Azadi and Emam Hussein Hospitals during 2010 and 2011 in Tehran, Iran. The Ethics Committees of Azadi and Emam Hossein Hospitals approved this study. The Diagnostic and Statistical Manual of Mental Disorders-IV (DSM-IV) (American Psychiatric Association 1994) rules were applied while diagnosing the patients. Medical and paramedical staff volunteers were used as the healthy group (n = 71, mean age 46.70 ± 11.716) who were free of any signs of neuropsychiatric disorders. Both patients and control groups were free from diabetes, dyslipidemia and cardiovascular disease. Consent forms were collected from all participants once the research had been thoroughly expressed. Demoghraphic characterization of the population is shown in Table 1.
| Variables | Cases | Controls | P Valuec |
|---|---|---|---|
| Age, y, mean ± SD | 47.53 ± 10.801 | 46.70 ± 11.716 | ˃ 0.05 |
| Age of onset | 20.79 ± 8.729 | - | - |
| Gender | |||
| Female | 26 | 29 | |
| Male | 67 | 70 | ˃ 0.05 |
| Smoking status | |||
| Non-smokers | - | 54 | - |
| Smokers | 93 | 45 | ˂ 0.001 |
| Educational level | |||
| Illiteracy | 3 | - | - |
| Primary | 8 | 1 | - |
| School | |||
| Guidance | 16 | 5 | - |
| High school | 53 | 28 | ˂ 0.001 |
| AD | 2 | 26 | - |
| BA | 8 | 28 | - |
| MA | 2 | 11 | - |
| Marital status | |||
| Single | 60 | 29 | - |
| Married | 17 | 64 | ˂ 0.001 |
| Divorced | 16 | 6 | - |
a Abbreviations: AD, associate diploma; BA, bachelor of arts; MA, master of arts
b Data are presented as No., otherwise indicated in the table.
c P values for Chi-square test.
3.2. DNA Isolation, DNA Modification, and Methylation-Specific PCR (MSP)
Two mL of blood was drawn from each participant for DNA extraction and further analysis (19). DNA bisulfate modification was prepared as earlier published (20, 21). Online software MatPrime was used to design methylated and unmethylated primers from the preferred sequences of APOE and CPT1-A promoters as shown in Table 2. Reaction for Methylation-specific PCR (MSP) was prepared in a total volume of 25 mL of liquid, including 16.0 mL double distilled water, Star Taq 5 U/mL,1 mL of dNTPs mix (10 mmol/L), 2 mL of Mg (25 mmol/L), and 0.5 mL of each primer (10mmol/L). MSP reactions for APOE gene were subjected to an initial incubation at 95ºC for 5 minutes, followed by 40 cycles of denaturation at 95˚C for 40 seconds, annealing at 58˚C for methyl primer and 59˚C for unmethyl primer for 30 seconds and extension at 72˚C for 30 seconds. Final extension was completed by incubation at 72˚C for 10 minutes. MSP reactions for CPT1-A gene were subjected to an initial incubation at 95ºC for 5 minutes, followed by 40 cycles of denaturation at 95˚C for 40 seconds, annealing at 58˚C for methyl primer and 62˚C unmethyl primer for 30 seconds and extension at 72˚C for 30 seconds. Final extension was completed by incubation at 72˚C for 10 minutes.
| Genes | Sequences (5´-3´) | Annealing Temperature (˚C) | Product Size |
|---|---|---|---|
| Cpt-1A M | F:TAGGAGGTATATCGACGGTTAAC | - | - |
| R:AAACAAATCTCTACTAAAAAACGAA | 58 | 184 | |
| Cpt-1A U | F:TAGGAGGTATATTGATGGTTAATGT | 62 | 184 |
| R:AAACAAATCTCTACTAAAAAACAAA | - | - | |
| APOE M | F:GTTGGGGTTAGTTGATGTTTATTAC | 58 | 203 |
| R:AAAAAAACTAAACTCCTAATTCGAA | - | - | |
| APOE U | F:GGTTGGGGTTAGTTGATGTTTATTAT | 59 | 204 |
| R:AAAAAAACTAAACTCCTAATTCAAA | - | - |
3.3. RNA Isolation and Real Time PCR
RNA isolation from fresh EDTA blood was conducted as earlier described (22). Revert Aid First-Strand cDNA Synthesis Kit (Fermentas, Cat. No. K1621) was used for reverse transcription of 1 mg RNA, according to manufacturer's instructions, in a final volume of 20 mL. AB15700 sequence detection system (Applied Biosystem) was utilized to analyze real time quantitative PCR of targeted cDNA. Amplification of each sample was performed in triplicate in a 25 µL reaction mixture containing 1 µL of CDNA, 10 µL of 2x_Sybergreen, 1 µL of each specific primer, and 8 µL of RNase free water. Primers which were used for expression analysis are indicated in Table 3.
| Genes | Sequences | Annealing Temperature |
|---|---|---|
| RNA 18s (Real Time- PCR) | 60°C | |
| F: GTAACCCGTTGAACCCCATT | ||
| R: CCATCCAATCGGTAGTAGCG | ||
| APOE (Real Time- PCR) | 60°C | |
| F: AGAGTTGGATGTGACCCTGCA | ||
| R: TCAGCTAAGGAGATGTGAGGACC | ||
| Cpt-1A (Real Time- PCR) | 60°C | |
| F: TCACCCTGTACCAGTCCAATACC | ||
| R: CTGCCTCCTGCTGGTCCTT T |
3.4. Statistical Analysis
Binary Logistic Regression Model was used for assessment of odds ratio (OR) and 95 % confidence intervals (95 % CI) of methylation status between cases and controls, while the Mann–Whitney test was used to analyze the expression level between cases and controls.
4. Results
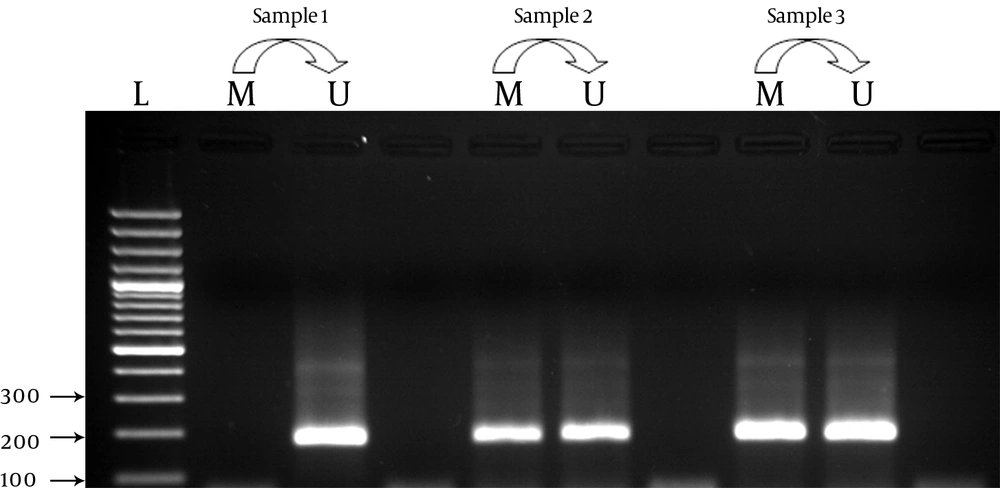
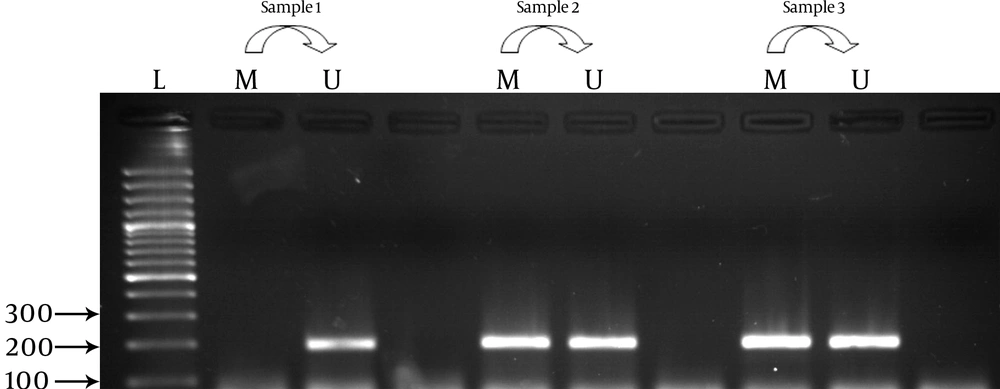
As shown in Table 2 methylation frequency for the APOE gene was 57 (71.25 %) for cases and 57 (80.28%) for controls. It was highlighted as a statistically insignificant difference between cases and controls regarding comparison of methylated pattern to unmethylate as reference. Analysis of the CPT1-A gene showed insignificant results in cases (43.75%: n = 35) compared to controls (40.84%: n = 29) concerning the status of promoter methylation(Table 4). Promoter methylation photographs are shown in Figure 1 and Figure 2. Gene expression profile of the cases and controls were not significant for the APOE and CPT1-A genes. As shown in Table 5, analysis of relative gene expression for APO E was (mean ± SD = 0.9252 ± 0.41611; Range = 0.35-1.75) and CPT1A was (mean ± SD = 0.4209 ± 0.37185; Range = 0.12-1.54) which was not significant.
| Genes | Methylation Status | Controls, No. (%) (n = 71) | Cases, No. (%) (n = 80) | P Value |
|---|---|---|---|---|
| APOE | 0.198 | |||
| Present | 57 (80.28) | 57 (71.25) | ||
| Absent | 14 (19.72) | 23 (28.75) | ||
| CPT1A | 0.718 | |||
| Present | 29 (40.84) | 35 (43.75) | ||
| Absent | 42 (59.15) | 45 (56.25) |
| Genes | Patients, No. | mean ± SD | Range | P Value |
|---|---|---|---|---|
| APOE | 0.174 | |||
| Cases | 17 | 0.9252 ± 0.41611 | 0.35 - 1.75 | |
| Controls | 17 | 1.1050 ± 0.32328 | 0.61 - 1.73 | |
| Cpt-1A | 0.986 | |||
| Cases | 17 | 0.4209 ± 0.37185 | 0.12 - 1.54 | |
| Controls | 17 | 0.4426 ± 0.33979 | 0.07 - 1.21 |
5. Discussion
Our results indicated that the promoter methylation variation of the APOE and Cpt1-A genes have no effect on the progress of SCZ and provide some initial evidence for APOE and CPT1-A genes in patients with SCZ. One of the main physiological processes influenced by different alleles of APOE is the sequestration of NMDA (N-methyl-D-aspartate) and AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionicacid) receptors in intracellular compartments, which could lead to changes in long-term potentiation (LTP) induction and neuronal degeneration. Therefore, this protein possibly plays a considerable role in synaptic plasticity by impairing the glutamatergic neurotransmission pathway (23, 24). Hasan et al. (2011) indicated that disturbed neuronal plasticity is considered to be part of the pathophysiology of schizophrenia and has been linked to the different clinical features of this severe illness (25). Concerning the role of APOE isoforms in the pathogenesis of schizophrenic disorders periodic controversial data has been published so far, certain of them highlight the significant role of APOE in development of SCZ (26, 27). Other studies indicated that the APOE gene is not of major importance in progress of SCZ (28-31). Daniel Martins-De-Souza et al. (2010) has found that expression analysis of apolipoprotein E in cerebro-spinal fluid may be a potential diagnostic tool for schizophrenia. In line with our study, Shinkai et al. (1998) have concluded that variation in the regulatory region of the APOE gene is not a risk factor for susceptibility to schizophrenia (32).The abnormal CpG island methylation of CPT1A took place during the differentiation of human embryonic stem cells (hESC) (33). Burke et al. (2006) have reported that environmental factors may contribute to the endophenotype of schizophrenia through the activation of specific genes such as CPT1. Prabakarans et al. (2004) showed that the transcriptional level of CPT1 significantly increases in brain of individuals with the SCZ disorder (18). The present study is the first evidence to highlight the position of promoter methylation of the APOE and CPT1 genes as a key regulator in CNS. Our outcomes may open a new window for studying the pathogenesis of schizophrenia, suggesting more studies with larger sample sizes in varied populations to validate this data.