1. Background
Pseudomonas aeruginosa is a gram-negative bacillus, an aerobic, non-fermenting, spore-free, motile bacterium with a capsule-like cell enamel layer on its surface. In nature, it thrives in moist environments and can even survive in distilled water (1). In hospital settings, it is commonly found in incubators, sinks, urinary catheters, and various medical supplies, as well as on employees' clothing and hands.
The rise in antibiotic resistance, particularly multidrug resistance, in this bacterium presents significant challenges, complicating infection treatment (2). Resistance in Pseudomonas aeruginosa arises through several mechanisms, including altered permeability to drugs, efflux pumps, receptor modification, access to secondary metabolic pathways, and the production of drug-degrading enzymes. Among these, the production of drug-degrading enzymes is a primary mechanism, actively degrading antibiotics and conferring resistance (3).
Pseudomonas aeruginosa has a strong preference for moist environments and is widely found in soil, water, plants, manure, and animal skin. Additionally, it may sometimes reside on the skin, in the external ear, upper respiratory tract, or large intestine of healthy individuals and can cause a range of diseases in humans and livestock. These include urinary tract infections, chronic pneumonia in cystic fibrosis patients, eye and ear infections, wound infections, and mastitis, often with antibiotic resistance (4).
Herbal remedies have long been used to treat various ailments, with approximately 45,000 known plant species (5). Nanotechnology, a rapidly growing field, currently impacts many areas of human life, including industry, agriculture, and medicine. The synthesis of metal nanoparticles has garnered significant interest due to their applications in scientific and industrial fields (6).
Among various nanoparticles, silver nanoparticles have demonstrated wide-ranging benefits in bio-nanotechnology and medicine (7). In medical science, silver nanoparticles are used for treating burns, producing dental materials, metal coatings, improving water purification, and as disinfectants. They also exhibit antifungal, antiviral, antibacterial, anti-inflammatory, anti-angiogenic, and anticancer activities (8).
Recently, biosynthesis using natural and biodegradable agents such as bacteria (9), fungi (10), and plants (11) has been established in various studies and is currently the primary focus of researchers in this field.
In the green synthesis method, metal ions are reduced to nanoparticles using plant compounds in a typically one-step reaction that does not require surfactants or other stabilizing agents. Bioactive substances and compounds in plant extracts, including flavonoids and other water-soluble metabolites, can reduce metal ions to nanoparticles at room temperature (12).
The antimicrobial activity of silver nanoparticles has been reported in numerous studies. The antibacterial mechanism of silver nanoparticles remains a topic of debate and is not yet fully understood. In studies on Escherichia coli and Staphylococcus aureus, it was observed that silver ions are released by nanoparticles, accumulate around the cell wall or within the cell, and impact DNA replication by interacting with protein thiol groups, causing protein inactivation (13). This interaction leads to the formation of reactive oxygen species (14).
Recent studies have demonstrated the antimicrobial, antioxidant, and anticancer activities of green-synthesized AgNPs (15-17). Biosynthesized silver nanoparticles have also been effectively used to degrade various chemicals (18).
Additionally, green-synthesized silver nanoparticles have a wide range of applications in biotechnology, including water filtration, disinfection, food preservation, cosmetics production, and the development of nano-insecticides and nano-pesticides (19-22).
The genus Capparis includes about 150 - 200 species found in tropical and subtropical regions worldwide (23), with four native species in Iran (24). In Iranian traditional medicine, Capparis species are used as anti-inflammatory, analgesic (for gout and rheumatoid arthritis), diuretic, antibacterial, and anthelmintic agents (25).
Several compounds, including alkaloids, terpenes, flavonoids, and fatty acids, have been reported from different Capparis species (26). Scientific studies have shown that these plants possess biological activities, including antioxidant, antibacterial, anti-glycemic, blood fat-lowering, and pain-relieving effects.
The herbal medicine market has surpassed $60 billion annually and continues to grow (27). Medicinal plants such as Capparis, which play a vital role in health systems, are therefore of considerable importance (27).
C. spinosa is one of the most economically significant species in the Capparidaceae family, with a vast diversity comprising approximately 40 - 50 genera and 700 - 900 species (28). Capparis is native to the Mediterranean basin and is widely distributed from Morocco to Crimea, Armenia, and Iran (29).
Several countries, including Greece, Italy, Spain, and Turkey, have extensive C. spinosa production (30). Spain and Turkey produce approximately 1,000 and 4,500 tons of C. spinosa (commonly known as snake grass) per year, respectively (31).
Afsharypuor et al. (32) reported that the leaves and ripe fruits of C. spinosa contain degradation products of glucosinolates, specifically methyl, isopropyl, and sec-butyl isothiocyanates.
2. Objectives
Glucosinolates are plant secondary metabolites of medicinal importance, as they may contribute to disease prevention and reduce the risk of carcinogenesis. Similarly, Çaliş et al. (33) reported the presence of indole-3-acetonitrile glycosides in C. spinosa fruits.
3. Methods
3.1. Preparation of Extract
Five grams of powder prepared from the snake grass plant was mixed with 100 cc of deionized water and heated for 15 minutes at boiling temperature. After cooling, it was filtered using Whatman No. 1 paper, and the resulting aqueous extract was stored at 4°C for subsequent tests.
3.2. Synthesis of Nanoparticles
Ten cc of the prepared extract was mixed with 90 cc of a 1 mM silver nitrate solution, and the mixture was placed on a magnetic stirrer for 24 hours at room temperature. To observe color changes, the solution's absorbance was measured using a spectrophotometer in the range of 300 - 700 nm. The solution containing the synthesized nanoparticles was then centrifuged at 1200 rpm for 15 minutes, after which the supernatant was discarded.
3.3. X-ray Diffraction Analysis
The X-ray diffraction (XRD) method was used to examine the metallic nature of the produced nanoparticles. After biological reduction, the silver nanoparticle solution was centrifuged at 8000 rpm for 5 minutes. The resulting pellet was re-dissolved in 10 mL of sterilized deionized water and centrifuged three times to purify it. The final pellet was then dried in a vacuum oven for 24 hours. The structure and composition of the purified nanoparticles were subsequently analyzed using XRD.
3.4. Fourier Transform Infrared Spectroscopy
The dried powder of the produced nanoparticles, along with the leaf extract powder, was mixed with KBr, formed into pellets, and analyzed using the device.
3.5. Isolation of Bacteria
The strains of Pseudomonas aeruginosa used in this research were collected and isolated from various surfaces in the maternity department of Amirul Mominin Hospital in Zabol city. To identify Pseudomonas aeruginosa, gram staining, catalase, and oxidase tests were conducted, along with confirmatory sugar tests using triple sugar iron agar (TSI).
Subsequently, the minimum inhibitory concentration (MIC) of all strains was determined for different groups of antibiotics, including cotrimoxazole, cephalexin, cefotaxime, cefazolin, gentamicin, imipenem, ampicillin, and ceftazidime, using the agar dilution method according to the clinical and laboratory standards institute (CLSI) guidelines. To determine the MIC, the standard reference powder for each of the antibiotics was used. In the first step, the required amount of each antibiotic for the initial dilution was dissolved in specific solvents. Following this, serial dilutions of each antibiotic were incorporated into Mueller Hinton agar culture medium. After bacterial culturing and 18 hours of incubation, the lowest concentration of each antibiotic at which no colony growth was observed was recorded as the MIC. Data were collected and analyzed using MS Excel 2010 software.
4. Results
The results of this study showed that the lowest inhibitory concentrations of the antibiotics ceftriaxone, cephalexin, cotrimoxazole, gentamicin, cefotaxime, imipenem, ceftazidime, ampicillin, and cefazolin were 128, 64, 32, 16, 4, 64, 64, 8, and 64 μg/mL, respectively. At these concentrations, 2, 3, 1, 2, 3, 2, 4, 2, and 3 strains of Pseudomonas aeruginosa were inhibited (Table 1).
| Bacterial | Ceftriaxone | Cephalexin | Cotrimoxazole | Gentamicin | Cefotaxime | Imi- Penin | Ceftazidime | Ampicillin | Cefazolin |
|---|---|---|---|---|---|---|---|---|---|
| 1. P. aeruginosa | 256 | 128 | 256 | 512 | 8 | 512 | 512 | 32 | 512 |
| 2. P. aeruginosa | 1024 | 256 | 128 | 128 | 8 | 1024 | 256 | 125 | 512 |
| 3. P. aeruginosa | 256 | 128 | 512 | 64 | 4 | 1024 | 64 | 64 | 512 |
| 4. P. aeruginosa | 256 | 64 | 512 | 64 | 4 | 1024 | 64 | 512 | 1024 |
| 5. P. aeruginosa | 1024 | 512 | 64 | 32 | 4 | 256 | 64 | 512 | 1024 |
| 6. P. aeruginosa | 512 | 512 | 64 | 128 | 16 | 256 | 64 | 256 | 512 |
| 7. P. aeruginosa | 512 | 1024 | 128 | 256 | 32 | 1024 | 256 | 128 | 512 |
| 8. P. aeruginosa | 512 | 512 | 128 | 256 | 32 | 1024 | 512 | 64 | 512 |
| 9. P. aeruginosa | 512 | 256 | 1024 | 32 | 64 | 1024 | 256 | 64 | 1024 |
| 10. P. aeruginosa | 256 | 64 | 256 | 128 | 8 | 256 | 512 | 16 | 512 |
| 11. P. aeruginosa | 256 | 64 | 256 | 256 | 16 | 256 | 512 | 16 | 512 |
| 12. P. aeruginosa | 512 | 256 | 128 | 512 | 64 | 1024 | 1024 | 128 | 512 |
| 13. P. aeruginosa | 512 | 128 | 128 | 32 | 32 | 128 | 256 | 256 | 256 |
| 14. P. aeruginosa | 128 | 1024 | 512 | 16 | 64 | 64 | 128 | 256 | 64 |
| 15. P. aeruginosa | 1024 | 1024 | 512 | 16 | 32 | 64 | 128 | 128 | 64 |
| 16. P. aeruginosa | 128 | 512 | 1024 | 128 | 64 | 512 | 1024 | 8 | 512 |
| 17. P. aeruginosa | 512 | 512 | 32 | 128 | 16 | 1024 | 512 | 8 | 64 |
Abbreviation: MIC, minimum inhibitory concentration.
This study showed that the lowest and highest inhibitory concentrations of the antibiotics ceftriaxone, cephalexin, cotrimoxazole, gentamicin, cefotaxime, imipenem, ceftazidime, ampicillin, and cefazolin were 256, 128, 64, 64, 8, 128, 16, and 128 μg/mL, and 1024, 1024, 1024, 1024, 128, 1024, 1024, 1024, and 1024 μg/mL, respectively. At these concentrations, 10, 8, 6, 2, 4, 10, 7, 2, and 13 strains of Pseudomonas aeruginosa were destroyed (Table 2).
| Bacterial number | Ceftriaxone | Cephalexin | Cotrimoxazole | Gentamicin | Cefotaxime | Imipenem | Ceftazidime | Ampicillin | Cefazolin |
|---|---|---|---|---|---|---|---|---|---|
| 1. P. aeruginosa | 512 | 256 | 512 | 1024 | 16 | 1024 | 1024 | 64 | 1024 |
| 2. P. aeruginosa | 1024 | 512 | 256 | 256 | 16 | 1024 | 512 | 256 | 1024 |
| 3. P. aeruginosa | 512 | 256 | 1024 | 128 | 8 | 1024 | 128 | 128 | 1024 |
| 4. P. aeruginosa | 512 | 128 | 1024 | 128 | 8 | 1024 | 128 | 1024 | 1024 |
| 5. P. aeruginosa | 1024 | 1024 | 128 | 64 | 8 | 512 | 128 | 1024 | 1024 |
| 6. P. aeruginosa | 1024 | 1024 | 128 | 256 | 32 | 512 | 128 | 512 | 1024 |
| 7. P. aeruginosa | 1024 | 1024 | 256 | 512 | 64 | 1024 | 512 | 256 | 1024 |
| 8. P. aeruginosa | 1024 | 1024 | 256 | 512 | 64 | 1024 | 1024 | 128 | 1024 |
| 9. P. aeruginosa | 1024 | 512 | 1024 | 64 | 128 | 1024 | 512 | 128 | 1024 |
| 10. P. aeruginosa | 512 | 128 | 512 | 256 | 16 | 512 | 1024 | 32 | 1024 |
| 11. P. aeruginosa | 512 | 128 | 512 | 512 | 32 | 512 | 1024 | 32 | 1024 |
| 12. P. aeruginosa | 1024 | 512 | 256 | 1024 | 128 | 1024 | 1024 | 256 | 1024 |
| 13. P. aeruginosa | 1024 | 256 | 256 | 64 | 64 | 256 | 512 | 512 | 512 |
| 14. P. aeruginosa | 256 | 1024 | 1024 | 32 | 128 | 128 | 256 | 512 | 128 |
| 15. P. aeruginosa | 1024 | 1024 | 1024 | 32 | 64 | 128 | 256 | 256 | 128 |
| 16. P. aeruginosa | 256 | 1024 | 1024 | 256 | 128 | 1024 | 1024 | 16 | 1024 |
| 17. P. aeruginosa | 1024 | 1024 | 64 | 256 | 32 | 1024 | 1024 | 16 | 128 |
Abbreviations: MBC, minimum bactericidal concentration.
4.1. The Results of Fourier Transform Infrared Spectrometer Studies Related to Silver Nanoparticles
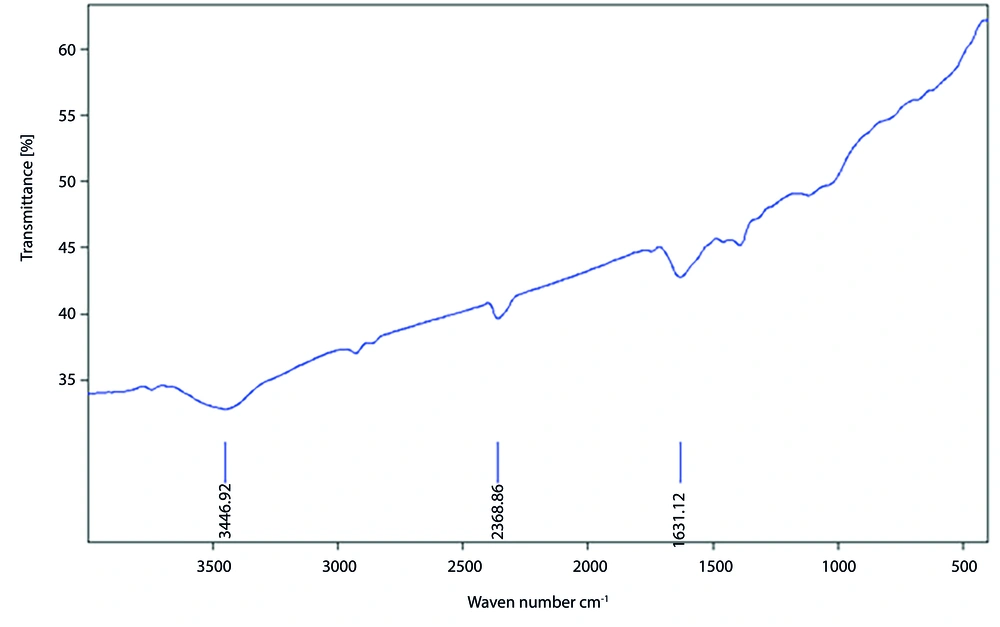
The results showed that the absorption peak observed in the range of 1680 - 1600 cm⁻¹ is related to the C=C stretching bond vibration, present in all studied samples, indicating the presence of alkene-containing compounds with covalent double bonds, such as ethene or ethylene. The absorption range of 2850 - 3000 cm⁻¹ corresponds to the presence of alkane compounds, such as methane and butane, in the sample. Additionally, the absorption observed in the range of 3600 - 3000 cm⁻¹ is associated with functional groups containing N-H and O-H bonds, suggesting the presence of phenolic compounds.
The vibration observed in the range of 1300 - 1400 cm⁻¹ corresponds to the N=O stretching bond, while the vibration in the range of 1400 - 1600 cm⁻¹ indicates the presence of aromatic compounds. The repeated observation of the absorption range of 2850 - 3000 cm⁻¹ further confirms the presence of alkane-containing chemical bonds, such as methane and butane, in the sample (Figure 1).
To examine the shape, size, and distribution of the nanoparticles, a transmission electron microscope (TEM) was used. The analysis revealed that the silver nanoparticles are spherical, highly dense, and well-dispersed, with a size of 150 nm in the stem extract.
The MIC of silver nanoparticles synthesized in medicinal plant extract was found to be 4 μg/mL, with 2 strains inhibited at this concentration. The maximum inhibitory concentration was 32 μg/mL, where 6 strains were inhibited (Table 3). The minimum and maximum lethal concentrations of the synthesized nanoparticles were 8 and 128 μg/mL, respectively, with 2 strains killed at each of these concentrations (Table 3).
| Bacterial Number | MIC | MBC |
|---|---|---|
| 1 | 8 | 16 |
| 2 | 4 | 8 |
| 3 | 4 | 8 |
| 4 | 8 | 16 |
| 5 | 16 | 32 |
| 6 | 16 | 32 |
| 7 | 32 | 64 |
| 8 | 8 | 16 |
| 9 | 8 | 16 |
| 10 | 32 | 64 |
| 11 | 32 | 64 |
| 12 | 64 | 128 |
| 13 | 64 | 128 |
| 14 | 32 | 64 |
| 15 | 32 | 64 |
| 16 | 16 | 32 |
| 17 | 32 | 64 |
Abbreviations: MIC, minimum inhibitory concentration, MBC, minimum bactericidal concentration.
5. Discussion
In this study, the MIC of antibiotics for Pseudomonas aeruginosa were as follows: Ceftriaxone, 128 µg/mL; cephalexin, 64 µg/mL; cotrimoxazole, 32 µg/mL; gentamicin, 16 µg/mL; cefotaxime, 4 µg/mL; imipenem, 64 µg/mL; ceftazidime, 64 µg/mL; ampicillin, 8 µg/mL; and cefazolin, 64 µg/mL.
Adabi et al. investigated the antibiotic resistance pattern of Pseudomonas aeruginosa in Zahedan. The results showed that beta-lactamase-producing strains were resistant to cefoxitin (96%), ceftazidime (15%), ceftriaxone (52%), cefotaxime (96%), cefotetan (30%), aztreonam (8%), and ciprofloxacin (8%) (34).
Habibi et al. studied the frequency and pattern of antibiotic resistance in Pseudomonas aeruginosa strains in Tehran hospitals. Their results showed that 88% of the strains were resistant to at least one antibiotic, with the highest resistance observed against cefotaxime (62.9%) and aztreonam (60.4%) (35).
In a study by Hemmati et al. examining Pseudomonas aeruginosa in Zanjan, the highest and lowest resistance rates were for cefotaxime (52 isolates, 43.3%) and amikacin (26 isolates, 21.6%), respectively. Resistance was also observed to imipenem in 35 isolates (29.2%), aztreonam in 45 isolates (37.5%), gentamicin in 45 isolates (37.5%), ceftazidime in 35 isolates (29.2%), and ciprofloxacin in 39 isolates (32.5%) (36).
Habibi et al. reported that, among 180 clinical samples, strains isolated from wound sources (44.1%) and urine (29.8%) had the highest frequency. Additionally, 97.5% of isolates exhibited beta hemolytic activity, while only 2.5% showed gamma hemolytic activity. Notably, 88% of isolates were resistant to one or more antibiotics (35).
In their study, Adabi et al. identified and confirmed strains using biochemical and genetic methods. Bacterial resistance to antibiotics was evaluated using the agar disk diffusion method, and MICs were determined for four representative antibiotics. Among 94 Pseudomonas aeruginosa strains, 83 (88.3%) exhibited multidrug resistance, with the highest resistance observed for cefepime (89.5%). For the antibiotics tested in MIC determinations, the highest resistance was to ciprofloxacin (89%) (37).
Jamshidi Gohar et al. found that Pseudomonas aeruginosa strains were most resistant to cephalothin, doxycycline, cefixime, amoxicillin, ampicillin, amikacin, and trimethoprim (38).
In the study by Rajabpour and Alikhani, the MIC of three different classes of antibiotics was investigated in Pseudomonas aeruginosa strains from hospitalized patients in Hamedan city. Among the eight selected antibiotics, the highest resistance was observed to ciprofloxacin, with 18 strains (58%) resistant, and levofloxacin, with 19 strains (61.2%) resistant. The lowest resistance was observed with imipenem, with only 3 strains (9.6%) resistant. Additionally, 2 strains (6.5%) showed complete resistance to all eight antibiotics. The MIC results showed that for gentamicin, 18 strains (58.06%) were resistant, 8 (25.08%) were intermediate, and 5 (16.12%) were sensitive. For ciprofloxacin, 28 strains (90.32%) were resistant and 3 (9.67%) intermediate. For imipenem, 12 strains (30.7%) were resistant, 13 (41.93%) intermediate, and 6 (19.35%) sensitive (39).
Golshani et al. investigated the resistance pattern of Pseudomonas aeruginosa strains in Isfahan hospitals. The resistance levels of the strains were as follows: Ciprofloxacin 56%, gentamicin 59%, tobramycin 61%, amikacin 65%, imipenem 55%, cefepime 55%, ceftazidime 57%, ceftriaxone 60%, cefotaxime 62%, oxacillin 100%, and piperacillin 48% (40).
In a study by Zarenia et al., the resistance pattern of Pseudomonas aeruginosa was investigated in samples from Kerman province. The results indicated the highest sensitivity was to cefizoxime, with 55 strains (91.7%), followed by imipenem (54 strains, 90%), meropenem (48 strains, 80%), and ciprofloxacin (40 strains, 66.7%). The highest resistance was observed with cefepime, with 36 strains (60%) resistant, and ciprofloxacin, with 19 strains (31.7%) resistant (41).
Roulova et al. studied the antibiotic resistance pattern of Pseudomonas aeruginosa isolated from six hospital wastewater sources. The resistance pattern, determined using the disc diffusion method for seven antibiotics, showed the highest resistance to ciprofloxacin (30.5%), followed by gentamicin (28.8%), meropenem (27.2%), ceftazidime (11.5%), amikacin (11.5%), piperacillin-tazobactam (11.5%), and aztreonam (8.5%) (42).
In the study by Zafer et al., the antibiotic resistance pattern of 122 Pseudomonas aeruginosa samples isolated from cancer patients was investigated. Resistance to beta-lactam antibiotics showed the following: Ceftazidime (60.6%), meropenem (45%), aztreonam (45.1%), imipenem (39.3%), and piperacillin/tazobactam (25.4%). Resistance to non-beta-lactam antibiotics included ciprofloxacin (43.4%), amikacin (32.18%), and polymyxin B (2.4%) (43).
In another study, 200 swab samples were obtained from the wound and burn department of an Iraqi teaching hospital. The results showed that out of 200 samples, only 31 were Pseudomonas aeruginosa. The antibiotic resistance pattern revealed resistance to gentamicin (83.87%), trimethoprim (67.74%), amikacin (54.83%), ceftazidime and tobramycin (25.80%), and bifloxacin and imipenem (22.58%) (44).
Jahantigh et al. investigated the antibiotic resistance pattern of Pseudomonas aeruginosa isolated from wounds at Zahedan Hospital. Their findings showed the highest resistance to trimethoprim/sulfamethoxazole (84.3%), cefipime (70.8%), piperacillin/tazobactam (20.8%), and colistin (8.3%) (45).
The antimicrobial properties of plant extracts are attributed to phenolic compounds and flavonoids, both of which are abundant in Capparis plant extracts (46, 47). Capparis extract is rich in phenolic, flavonoid, rutin, tocopherol, carotenoid, and vitamin C compounds (31, 48). Other studies have also demonstrated that Capparis extract is high in phenolic and flavonoid content (49).
Krishna Raj reported the minimum inhibitory concentration of nanosilver against Escherichia coli and Pseudomonas aeruginosa as 10 and 20 µg/mL, respectively, whereas in the present study, these values were 5 and 10 µg/mL, respectively (50).
In the study by Azizian Shermeh et al., silver nanoparticles were biosynthesized using the aqueous extract of mace plant leaves, and their antimicrobial activity was evaluated. Upon adding the extract to a silver nitrate solution, the solution turned brown. The silver nanoparticles showed maximum absorption at 405 nm, had a spherical shape, and ranged in size from 8 - 12 nm. These nanoparticles exhibited significant antimicrobial activity against tested samples, inhibiting bacterial and fungal growth at very low concentrations (51).
In 2014, Shams et al. investigated the antimicrobial properties of silver nanoparticles derived from lentil seed extract against several gram-positive and gram-negative bacteria. The study confirmed that silver nanoparticles are effective bactericidal agents against a wide range of gram-positive and gram-negative bacteria, including antibiotic-resistant strains (52).
5.1. Conclusions
The results of this study showed that synthesized silver nanoparticles demonstrated a strong inhibitory effect on antibiotic-resistant Pseudomonas aeruginosa. Given that this bacterium has contributed to increased hospital infections, these nanoparticles could potentially serve as a solution to reduce and eliminate bacterial strains.