1. Background
The honey bee is an insect belonging to the class of insects and the order of diptera, found worldwide across various geographical regions (1). Based on morphological and molecular studies, 26 bee subspecies have been identified, which fall into five evolutionary lines: The Northern and Western European line (M), Central and Southern Africa (A), the Northern Mediterranean region and Eastern Europe (C), the Eastern Mediterranean, Near East, and Eastern Middle East (O), and finally the Eastern group of Ethiopia (Y) (2, 3). Among the eight subspecies of bees living in the Middle East, one subspecies, Apis mellifera meda, is found in Iran, known as the Iranian honey bee (1). In addition to Iran, this subspecies also occurs in northern Iraq, southeastern Turkey, and northern Syria (2, 3). Morphologically, it is similar to the Italian breed. In terms of biological characteristics, it is a breed with a high reproductive capacity, the ability to collect propolis, excellent wintering ability, and high aggression, all of which distinguish it from other subspecies in the region (4).
To study different populations of organisms, several methods have been widely adopted by systematic researchers, one of which is genetic diversity (5). Genetic diversity is a crucial element for genetic progress, preservation, evolution, and adaptation to changing and diverse environmental conditions (6). Phenotypic flexibility and genetic variability in traits used to identify species can lead to misidentification. However, the use of more accurate and faster systems for identifying living organisms, including molecular identifiers, can aid in the process of sample identification (7). Among molecular markers, the mitochondrial and nuclear genomes are both very useful in studying evolution and resolving relationships between different animal populations (6). The mitochondrial genome is often preferred over the nuclear genome in animals due to its multiple copies in cells, low recombination rate, haploid inheritance, and ease of extraction (8).
One of the most suitable mitochondrial genes for research using animal molecular markers is the mitochondrial cytochrome oxidase I (COI) gene (cytochrome oxidase c subunit one), which has evolved in such a way that it can distinguish even very closely related species (9). Cytochrome oxidase is the final enzyme in the mitochondrial respiratory chain, responsible for transferring electrons from cytochrome to oxygen. It is a protein complex found in the inner mitochondrial membrane, composed of 13 protein subunits, 10 of which are encoded by the nuclear genome and the remaining 3 by the mitochondrial genome (10). Cytochrome c oxidase is an enzyme present in all living organisms (11). This fact suggests that the enzyme has ancient origins, likely first arising in a primitive organism and subsequently being passed down to all organisms during evolution across generations (12).
In honey bees, genetic studies are primarily based on the analysis of both morphological (13) and molecular data (14). These two methods are currently used as powerful tools to estimate genetic diversity and determine phylogenetic relationships between different populations of bee subspecies worldwide. While morphological characteristics are useful for phylogenetic studies, they are highly sensitive to environmental factors and changing environmental conditions. In contrast, for phylogenetic studies, enzyme digestion analysis using mitochondrial DNA information provides a better and more reliable genetic marker (15). One advantage of mitochondrial DNA over morphological data is that mitochondrial DNA is more easily analyzed in the context of phylogenetic issues. Additionally, due to maternal inheritance, lack of recombination, and low efficiency of complete repair mechanisms, mtDNA evolves more rapidly than single-copy genes in nuclear DNA. Therefore, mtDNA is particularly useful for studying phylogenetic relationships and the historical changes among bee subspecies (16).
2. Objectives
In general, the first step in achieving targeted breeding is to determine the level of genetic diversity within populations. Based on the aforementioned content, and in order to identify genetic reserves and preserve breed diversity, the present study was conducted to investigate the genetic and phylogenetic structure of native Iranian honey bees and several important honey bee species worldwide.
3. Methods
3.1. Identification and Editing of Cytochrome Oxidase I Gene Sequences
To identify COI gene sequences, the DNA and mRNA nucleotide sequences for the COI gene in honey bees were identified. Additionally, using the BLAST tool in genome databases, the COI gene sequences for other target species were obtained (Table 1). To compare the sequences and determine genetic parameters, the COI gene sequence of the Iranian honey bee was aligned with those of other species using Clustal W.
3.2. Phylogeny and Determining the Evolutionary Path
The phylogenetic tree of the COI sequence was constructed for the studied species using MEGA software. After editing the sequences and removing non-homogeneous regions, the phylogenetic tree was drawn using the Neighbor Joining (NJ) method. In this method, the matrix (Q) is used, which includes all the branches, and selects the lowest value that represents the highest similarity between two branches, which is then used in the phylogeny tree. Bootstrapping values were obtained by resampling 1,000 times.
3.3. Molecular Diversity Indices
Molecular diversity indices such as the number of polymorphic sites, number of haplotypes, haplotype frequency, nucleotide diversity, total number of mutations, nucleotide differences between populations, and indices of conservation in protected areas, such as sequence conservation, minimum conservation length, conservation threshold limit, and neutrality statistics, were analyzed using Dnasp software.
3.3.1. Molecular Diversity Indices
3.3.1.1. Polymorphic Sites
These are positions in the DNA sequence where variations occur among individuals in a population. A higher number of polymorphic sites indicates greater genetic diversity.
3.3.1.2. Haplotypes and Haplotype Frequency
Haplotypes refer to a combination of alleles at multiple loci that are transmitted together on the same chromosome. Haplotype frequency is the proportion of a specific haplotype relative to the total number of haplotypes in the population, providing insights into genetic structure.
3.3.1.3. Nucleotide Diversity (π)
This measures the average number of nucleotide differences per site between two sequences. It is a crucial metric for assessing genetic variation within a population.
3.3.1.4. Total Number of Mutations
This refers to the cumulative count of mutations observed in a given sequence, which can signify evolutionary changes over time.
3.3.1.5. Nucleotide Difference Between Populations
This index quantifies the genetic distance between different populations, helping to understand evolutionary relationships.
3.3.1.6. Conservation Indices
Sequence conservation: This measures how much of a sequence remains unchanged across different species or populations, indicating evolutionary pressure.
Minimum conservation length and conservation threshold limit: These metrics help establish how much of a sequence must be conserved for it to be considered functionally important.
Neutrality Statistics: These tests (e.g., Tajima's D) assess whether allele frequencies in a population conform to expectations under neutral evolution, which can indicate past demographic events.
| No. | Scientific Name | ID Number in the NCBI Database |
|---|---|---|
| 1 | Apis andreniformis Aan 4.1 | LC427579.1 |
| 2 | Apis andreniformis voucher AAT95 | MT670338.1 |
| 3 | Apis andreniformis voucher AAT97 | MT670340.1 |
| 4 | Apis cerana isolate 8 | OK253002.1 |
| 5 | Apis cerana indica voucher ACIOR1 | OP168349.1 |
| 6 | Apis cerana indica breed Honey bee | MW093739.1 |
| 7 | Apis koschevnikovi Ak1_TB | LC461206.1 |
| 8 | Apis koschevnikovi Ak1_HST | LC461204.1 |
| 9 | Apis koschevnikovi Ak1_BL | LC461205.1 |
| 10 | Apis florea voucher UBN3 | OK350423.1 |
| 11 | Apis florea voucher UBN1 | OK350421.1 |
| 12 | Apis florea voucher SMI9 | OK350397.1 |
| 13 | Apis dorsata isolate Jatoli | OQ346377.1 |
| 14 | Apis laboriosa voucher ALM6 | MT679379.1 |
| 15 | Apis laboriosa voucher ALM5 | MT679378.1 |
| 16 | Apis laboriosa voucher ALM1 | MT679374.1 |
| 17 | Apis nigrocincta AnBK4.2 | LC534595.1 |
| 18 | Apis nigrocincta AnBK3.3 | LC534593.1 |
| 19 | Apis nigrocincta AnBK3.2 | LC534592.1 |
| 20 | Apis mellifera voucher HY42 | OP435367.1 |
| 21 | Apis mellifera voucher HY41 | OP435365.1 |
| 22 | Apis mellifera isolate mellifera | ON680902.1 |
| 23 | Apis mellifera isolate ACKU | OM320635.1 |
| 24 | Apis mellifera meda isolate DUHOK29-9 | OP020350.1 |
| 25 | Apis mellifera meda isolate DUHOK29-7 | OP020348.1 |
| 26 | Apis mellifera meda isolate DUHOK29-6 | OP020347.1 |
4. Results
4.1. Experiment and Analysis of Genetic Distance and Phylogenetic Tree
The results of the evolutionary diversity in the 26 studied species are presented in Table 2. Since the genetic distances between different species are calculated pairwise, the resulting numbers represent the nucleotide substitutions between the studied sequences. The genetic distance between two species is related to their phylogenetic correlation, making this index useful for determining the genetic distance and proximity between species.
| Variables | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Specie 1 | |||||||||||||||||||||||||
| Specie 2 | 1.293 | ||||||||||||||||||||||||
| Specie 3 | 1.293 | 0.000 | |||||||||||||||||||||||
| Specie 4 | 0.119 | 1.712 | 1.712 | ||||||||||||||||||||||
| Specie 5 | 1.221 | 0.124 | 0.124 | 1.168 | |||||||||||||||||||||
| Specie 6 | 1.221 | 0.129 | 0.129 | 1.482 | 0.013 | ||||||||||||||||||||
| Specie 7 | 1.173 | 0.123 | 0.123 | 1.302 | 0.114 | 0.098 | |||||||||||||||||||
| Specie 8 | 1.173 | 0.123 | 0.123 | 1.302 | 0.114 | 0.098 | 0.000 | ||||||||||||||||||
| Specie 9 | 1.162 | 0.119 | 0.119 | 1.285 | 0.111 | 0.098 | 0.003 | 0.003 | |||||||||||||||||
| Specie 10 | 1.372 | 0.096 | 0.096 | 1.483 | 0.108 | 0.119 | 0.114 | 0.114 | 0.111 | ||||||||||||||||
| Specie 11 | 1.372 | 0.096 | 0.096 | 1.483 | 0.108 | 0.119 | 0.114 | 0.114 | 0.111 | 0.000 | |||||||||||||||
| Specie 12 | 1.370 | 0.094 | 0.094 | 1.480 | 0.105 | 0.117 | 0.111 | 0.111 | 0.108 | 0.002 | 0.002 | ||||||||||||||
| Specie 13 | 1.306 | 0.120 | 0.120 | 1.390 | 0.117 | 0.103 | 0.124 | 0.124 | 0.127 | 0.134 | 0.134 | 0.132 | |||||||||||||
| Specie 14 | 1.233 | 0.115 | 0.115 | 1.560 | 0.100 | 0.090 | 0.102 | 0.102 | 0.105 | 0.102 | 0.102 | 0.100 | 0.103 | ||||||||||||
| Specie 15 | 1.233 | 0.115 | 0.115 | 1.560 | 0.100 | 0.090 | 0.102 | 0.102 | 0.105 | 0.102 | 0.102 | 0.100 | 0.103 | 0.000 | |||||||||||
| Specie 16 | 1.233 | 0.115 | 0.115 | 1.560 | 0.100 | 0.090 | 0.105 | 0.105 | 0.105 | 0.102 | 0.102 | 0.100 | 0.106 | 0.002 | 0.002 | ||||||||||
| Specie 17 | 0.113 | 1.253 | 1.253 | 0.074 | 1.240 | 1.167 | 1.138 | 1.138 | 1.128 | 1.237 | 1.237 | 1.234 | 1.288 | 1.133 | 1.133 | 1.133 | |||||||||
| Specie 18 | 0.112 | 1.239 | 1.239 | 0.068 | 1.225 | 1.153 | 1.125 | 1.125 | 1.115 | 1.222 | 1.222 | 1.220 | 1.273 | 1.119 | 1.119 | 1.119 | 0.008 | ||||||||
| Specie 19 | 0.112 | 1.239 | 1.239 | 0.068 | 1.225 | 1.153 | 1.125 | 1.125 | 1.115 | 1.222 | 1.222 | 1.220 | 1.273 | 1.119 | 1.119 | 1.119 | 0.008 | 0.000 | |||||||
| Specie 20 | 1.315 | 0.142 | 0.142 | 1.570 | 0.127 | 0.125 | 0.117 | 0.117 | 0.120 | 0.120 | 0.120 | 0.118 | 0.105 | 0.103 | 0.103 | 0.105 | 1.187 | 1.173 | 1.173 | ||||||
| Specie 21 | 1.310 | 0.144 | 0.144 | 1.576 | 0.129 | 0.125 | 0.117 | 0.117 | 0.121 | 0.122 | 0.122 | 0.120 | 0.108 | 0.105 | 0.105 | 0.108 | 1.187 | 1.173 | 1.173 | 0.002 | |||||
| Specie 22 | 0.116 | 1.733 | 1.733 | 0.095 | 1.309 | 1.633 | 1.359 | 1.359 | 1.342 | 1.581 | 1.581 | 1.579 | 1.388 | 1.603 | 1.603 | 1.603 | 0.112 | 0.106 | 0.106 | 1.592 | 1.597 | ||||
| Specie 23 | 1.311 | 0.147 | 0.147 | 1.711 | 0.127 | 0.125 | 0.113 | 0.113 | 0.115 | 0.119 | 0.119 | 0.117 | 0.105 | 0.107 | 0.107 | 0.109 | 1.187 | 1.173 | 1.173 | 0.000 | 0.002 | 1.751 | |||
| Specie 24 | 0.113 | 1.575 | 1.575 | 0.108 | 1.325 | 1.531 | 1.398 | 1.398 | 1.386 | 1.519 | 1.519 | 1.517 | 1.372 | 1.481 | 1.481 | 1.481 | 0.108 | 0.104 | 0.104 | 1.453 | 1.445 | 0.012 | 1.615 | ||
| Specie 25 | 0.113 | 1.575 | 1.575 | 0.108 | 1.325 | 1.531 | 1.398 | 1.398 | 1.386 | 1.519 | 1.519 | 1.517 | 1.372 | 1.481 | 1.481 | 1.481 | 0.108 | 0.104 | 0.104 | 1.453 | 1.445 | 0.012 | 1.615 | 0.000 | |
| Specie 26 | 0.113 | 1.575 | 1.575 | 0.108 | 1.325 | 1.531 | 1.353 | 1.353 | 1.341 | 1.519 | 1.519 | 1.517 | 1.372 | 1.481 | 1.481 | 1.481 | 0.108 | 0.104 | 0.104 | 1.453 | 1.445 | 0.012 | 1.615 | 0.000 | 0.000 |
Based on the Poisson correlation of the evolutionary matrix, the calculation and results indicated that the Iranian honey bee exhibits the highest genetic distance (1.615) with the A. mellifera isolate ACKU species, and the lowest genetic distance (0.012) with the A. mellifera isolate Mellifera species. The numbers obtained from the genetic distance matrix reflect the degree of nucleotide substitution between the sequences. The genetic distance between two species is closely related to their phylogenetic correlation, making this index useful for assessing the distance or proximity between species (17).
Phylogenetic analysis estimates how members of a family may have derived from one another during the course of evolution. These relationships are determined by studying mutations such as substitutions, deletions, insertions, and rearrangements that are subject to natural selection. The purpose of phylogenetic analysis is to uncover a branching arrangement in the form of a tree that illustrates the best relationships between sequences. It also enables the discovery of gene functions, the tracing of gene origins, and the identification of an organism's relatives (17).
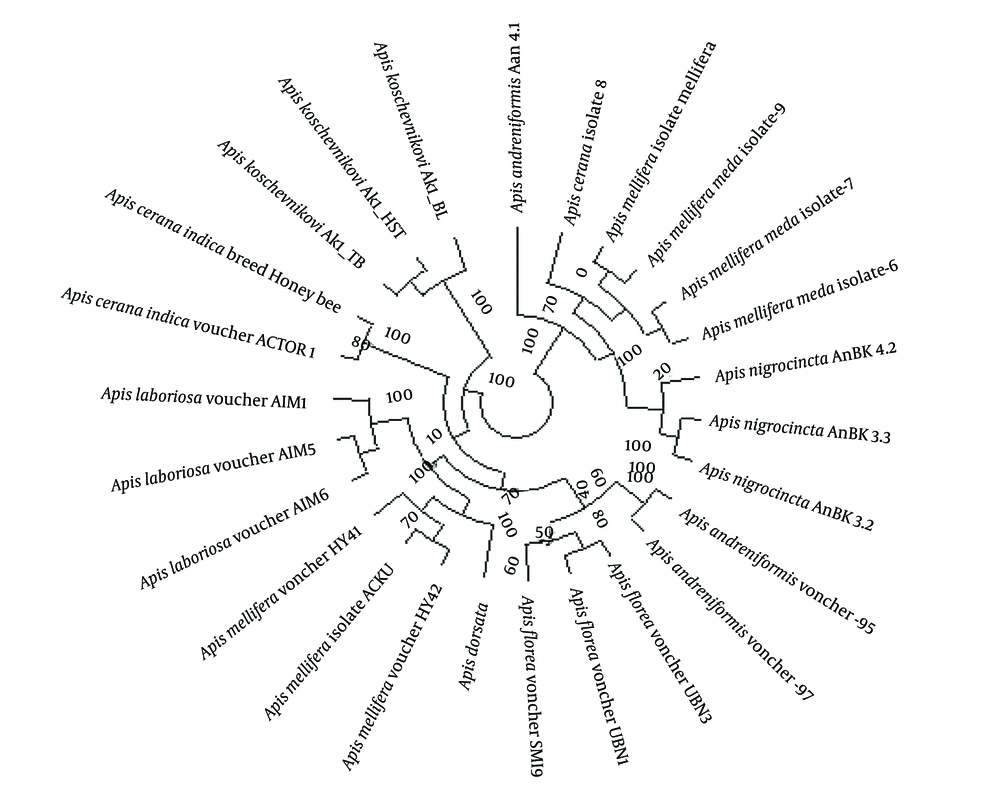
Evolutionary relationships between sequences are represented using diagrams called phylogenetic trees. The purpose of phylogenetic analysis is to discover an arrangement of branches in the form of trees that show the best relationships of sequences. Based on gene sequence cluster analysis, the studied species were classified into nine categories (Figure 1). Category one is related to Apis koschevnikovi; the second category is related to Apis cerana indica; the third category is related to Apis laboriosa; the fourth category is related to Apis dorsata and Apis mellifera species (excluding the Mellifera subspecies); the fifth category is related to Apis andreniformis species (excluding the Ana 4.1 subspecies) and Apis flora; the sixth category is related to Apis nigrocincta; the seventh category is related to Apis mellifera meda and Mellifera Apis mellifera isolate species; and the eighth and ninth categories are related to Apis cerana isolate 8 and Apis andreniformis Ana 4.1 species, respectively. The highest and lowest phylogenetic distances for the Iranian honey bee are with Apis koschevnikovi and Mellifera Apis mellifera isolate, respectively.
4.2. Nucleotide Substitutions
The results of the substitution percentages for COI gene nucleotide sequences, shown in Table 3, indicate that the highest substitution values are associated with pyrimidine bases. Specifically, the conversion of thymine to cytosine was 9.8%, while the conversion of cytosine to thymine was 31.51%. These values were lower for purine bases, with the conversion of adenine to guanine and guanine to adenine at 2.74% and 7.84%, respectively.
a This table shows the percentage of substitution from a row to a column.
b The numbers outside the diagonal represent crossover substitutions.
c The numbers on the diagonal represent transition substitutions.
4.3. Replacement Rate
Based on the results in Table 2, the dN/dS ratio for the COI gene was calculated for different bee species, with the numerical value of the ratio (dN/dS) being 0.08. If this ratio is greater than one, it indicates positive selection; if it is less than one, it indicates purifying selection; and if it is equal to one, it indicates neutral selection during gene evolution (18, 19). In this study, the dN/dS ratio was found to be less than one, suggesting purifying selection during the evolution of the COI gene (Table 4).
| Parameters | Numerical Value | Standard Deviation |
|---|---|---|
| dN | 0.030950 | 0.008807 |
| dS | 0.386167 | 0.041641 |
| dN/dS | 0.080146 | 0.000154 |
Abbreviations: dN, nucleotide changes that cause amino acid changes; dS, nucleotide changes that do not affect the resulting amino acid; of dN/dS the process of natural selection.
4.4. Genetic Diversity Indices
The nucleotide diversity and haplotype diversity in terms of base pairs for the studied bee populations were estimated at 0.259 and 0.966, respectively. The average nucleotide difference between ecotypes was calculated as 83.19 bp. Additionally, in the analysis of genetic diversity indices for the COI region of the mitochondrial genome in the native Iranian bee ecotype, compared with the nucleotide sequences of this region in the other studied bee ecotypes, 20 point mutations were identified. These mutations were distributed differently along the length of the mitochondrial genome. In research conducted in Pakistan and China, the COI region of the mitochondrial genome (20) was also used to study the genetic structure of the honey bee (A. mellifera) population. The results indicated that, in the entire studied population, 44 polymorphisms, 2.429 inter-population nucleotide differences, and 23 mutations were detected (Table 5).
| Indices | Theta-W | S | H | Hd | Pi | Eta | K |
|---|---|---|---|---|---|---|---|
| Values | 0.159 | 195 | 4 | 0.966 | 0.259 | 20 | 83.19 |
Abbreviations: Theta-W, mutation rate calculated from all polymorphic sites; S, number of polymorphic loci; H, number of haplotypes; Hd, haplotype frequency; Pi, nucleotide diversity; Eta, total number of mutations; K, nucleotide difference between populations or species (nucleotide divergence).
According to Table 6, the analysis of indices for the protected areas of the COI region of the mitochondrial genome in the studied population revealed the following values: Minimum protection threshold = 0.396, protection length = 80, and sequence protection = 0.49. Neutrality tests, including the Tajima's D test, were performed to identify any deviation from the null hypothesis of neutral evolution and to examine the effects of natural selection on genes across different populations. The numerical value for Tajima's D was estimated to be 1.174 in this study.
| Indices | D Tajima Test | CT | MWL | C |
|---|---|---|---|---|
| Values | 1.174 | 0.49 | 80 | 0.396 |
Abbreviations: CT, protection threshold limit; MWL, minimum protection length; C, sequence protection.
5. Discussion
Previous research on animals and various genes has concluded that the cytochrome oxidase gene is a useful and reliable marker for differentiating populations (21-23). In one study (24), COI genes were used to investigate the phylogenetic characteristics of honey bee populations. The results demonstrated the suitability of the COI gene for studying and differentiating various honey bee populations in Iran. Another study conducted by previous researchers (25) in Thailand used genetic comparisons of mitochondrial genes COI and COII to differentiate populations of the small honey bee (A. florea) and the small honey bee (A. andreniformis). The results showed that these genes are effective in distinguishing the populations of these two species. The mitochondrial COI gene was also employed in research to differentiate populations of bees collected from the northwestern Himalayan region, where the results indicated that the gene region plays an important role in separating populations and subspecies of honey bees in this area (26). In another study by previous researchers (27), the COI mitochondrial gene was used to distinguish different populations of the Apis genus. The findings confirmed that the COI gene, as a molecular marker, can be effectively used to analyze the ancestral status and evolution of Apis species and has a superior ability to differentiate between bee species.
In this study, the investigation of phylogenetic relationships showed that the genetic diversity of the evaluated ecotypes does not align with their geographical diversity. Although, in most cases, the study populations were classified based on their geographical origin, it was observed that, in some instances, two populations from geographically close areas were placed into completely separate groups in the population classification based on molecular data. For example, the ecotype of Apis mellifera meda from Iran and the ecotype of Apis andreniformis from Southeast Asia were classified into separate groups in the cluster analysis, despite both populations belonging to similar climates. Such results are also evident in the cluster analysis of previous research (24), the grouping of small honey bee populations using COI and COII mitochondrial genes in earlier studies (2, 20), the grouping of different Iranian honey bee populations using the mitochondrial ND2 gene in previous research (1), the study of the genetic diversity of bee populations in Kerman province using ISSR markers (16), and the investigation of phylogenetic relationships in Iranian honeybees using both morphological and molecular markers.
To explain cases of inconsistency between groupings based on molecular data and geographical origin, it can be suggested that these groupings may reflect the unique genetic backgrounds of these populations and the presence of specific features in their genome sequences. Additionally, placing populations from geographically distant areas into the same group based on molecular data may be due to factors such as germplasm transfer, the insufficient number of markers used in genetic diversity studies, or the broad distribution of a plant species within the studied geographical area. In other words, the geographical distance between populations cannot be considered conclusive evidence of the distance or proximity of their genetic backgrounds (28-30).
Four microsatellite loci of honey bee populations in northwestern Iran have been investigated in various studies, including the study of morphometric characteristics of honey bee (Apis mellifera L.) populations in Kermanshah province (16, 28, 31), the study of morphological diversity of the small honey bee in Iran using statistical methods such as cluster analysis and minimum variance (32), and an investigation of mitochondrial genes ND1 and ND5 to explore the separation of Iranian honey bees from global subspecies (33). Additionally, the genetic diversity of bee populations in Southeast Europe has been studied using 22 microsatellite markers (34), and the genetic diversity of honey bee populations in the Tomsk region of Russia was analyzed using COI and COII genes (35). Furthermore, the genetic diversity of Argentine honey bee populations has been examined using COI and COII genes (36). All these studies have shown that there is significant genetic diversity within the species, with notable intraspecies diversity at the DNA level. According to these researchers, factors such as colony migration, migratory beekeeping, and the random mating of queens with males from different colonies play important roles in creating diversity within honey bee populations.
The substitution values obtained in this study are consistent with those reported by other researchers, who found the highest substitution rate to be in pyrimidine bases (37). Generally, the cause of these changes can be attributed to cytosine methylation (38).
The dN/dS method is one of the key indicators used to detect the process of natural selection during gene evolution. In this method, the results of nucleotide changes that lead to amino acid changes (dN) are compared to nucleotide changes that do not affect amino acids (dS) (18, 19).
Populations that have undergone expansion, or experienced a significant increase in size, or that have been subject to directional selection, typically show negative and significant D values. On the other hand, positive and significant values of D indicate the effects of genetic drift, genetic bottlenecks, or balancing selection during the evolutionary history of the population. Genetic drift leads to increased homogeneity and the loss of alleles, particularly recessive alleles. Ultimately, these factors result in reduced diversity and a decreased ability of populations to adapt to environmental changes, a process that is especially common in smaller and more homogeneous populations (18, 39).
5.1. Conclusions
In conclusion, the results of this study indicate that the conserved region of the mitochondrial cytochrome oxidase gene among the species studied represents only a small portion of the entire sequence. This suggests a high level of polymorphism in this gene, as well as its susceptibility to nucleotide changes and mutations. A total of 20 mutations were identified during the evolution of this gene locus. Additionally, 195 polymorphic sites were observed, supporting the idea of purifying selection acting on this gene. This may play a crucial role in understanding the biological function of the gene, as it may lead to the emergence of new proteins and functions. Finally, by comparing the nucleotide sequences of Iranian honey bee populations with those of other honey bee populations from different regions of the world in gene banks, it was determined that the COI region is capable of distinguishing Iranian honey bee populations from other populations.
The study provides significant insights into the genetic diversity and phylogenetic relationships among various bee species, particularly the Iranian honey bee. The analysis reveals a notable genetic distance between the Iranian honey bee and other species, indicating its unique evolutionary path. The findings highlight the importance of mitochondrial COI gene sequences in understanding genetic variation and natural selection processes. Furthermore, the results underscore the necessity for ongoing research to preserve bee biodiversity and inform effective conservation strategies. Overall, this research contributes valuable data to the field of apidology and enhances our understanding of honey bee evolution.