1. Background
Staphylococcus species are one of the most common causes of nosocomial infections in the world (1). The incidence of staphylococcal infections in recent years has been increasing due to the release of resistant strains, increasing patients with immune deficiency and overuse of medical devices such as catheters (2).
Among Staphylococcus species, Staphylococcus aureus has the highest pathogenicity. Staphylococcus aureus is responsible for diseases including skin flashing syndrome, urinary tract infections, bacteremia, and toxic shock syndrome (3). The ability of Staphylococcus aureus isolates to produce the disease depends on the production of several different types of extracellular toxin. Most strains of S. aureus isolated from patients with symptoms of toxic shock syndrome (TSS) produce a toxin called toxin shock syndrome (TSST-1), which is a super antigen (4). Although classically, the cause of the disease was due to the use of tampon, however, many other things, such as stinging, could also cause the disease (5). Several different enterotoxins from S. aureus, all of which are super antigens (SEE-SEC-SEB-SEA-SHE-SEE), have been identified so far that, in addition to gastrointestinal poisoning, are also involved in toxic shock syndrome (6). SEC-SEB-SEA-SEI-SHE-SEJ toxins can cause symptoms of toxic shock syndrome (7). Staphylococcal enterotoxins are single-chain, low molecular weight proteins that are similar in composition and biological activity, but different in antigens. Staphylococcal enterotoxin-encoding genes are encrypted by mobile genetic elements such as bacteriophages (sep-see-sea) or plasmids (set-ses-ser-sej-sed), or by pathogenic islets on staphylococcal chromosomes (seb-sec- she-sei-sek-sel-sem-seq-sep-seo-sen) (8). Therefore, it can be said that enterotoxin can be transmitted horizontally between Staphylococcus strains and it is likely that these genetic elements play an important role in the development of S. aureus as a pathogen (9). Due to the need for the expression of enterotoxin genes in phenotypic techniques and the anti-genetic similarity of enterotoxin serotypes, there is a potential for cross-reactivity. Therefore, using genotypic methods to detect enterotoxin gene in S.aureus strains is preferred in clinical specimens (10). So far, most studies on toxins responsible for S. aureus syndrome have been carried out on food and animal resources (11). Therefore, there is a need for clinical information in human specimens in Iran and neighboring countries. Identification of toxin strains of S. aureus by immunological methods such as immunodiffusion, agglutination, radioimmunoassay, and ELISA is time consuming, difficult, and non-specific (12).
According to the studies, the PCR method, which is based on DNA replication, is a suitable tool for the rapid, sensitive, and specific detection of S. aureus toxin genes (13).
Type C enterotoxin is one of the 5 most common enterotoxins that interfere with food poisoning and toxic shock syndrome. It has been shown that enterotoxin C has contributed to the increase in the severity of the disease in the experimental infectivity of the animal model. This toxin has 3 biotypes (C1-C2-C3). It is believed that the S. aureus enterotoxin encoding gene is located on a plasmid containing an antibiotic resistance gene (14).
Some enterotoxin-C producing strains simultaneously produce other enterotoxins as well as toxins that cause toxic shock syndrome (15). Importantly, enterotoxins are not produced by S. aureus coagulase alone; however, in recent years it has been shown that negative Staphylococcus coagulase can also produce enterotoxins (16).
Enterotoxin E is a simple protein, spongy, white, and soluble in saline liquids. It has an antigenic property whose precursor molecular weight after final processing, using a saturation centrifuge and sucrose concentration of G-75 29300 to 30030 Dalton, is 26425 Dalton. Staphylococcal enterotoxin encoding genes encode plasmids on the sae’s chromosome fragments. The aim of this study is identification of the Staphylococcus aureus, types C and E, producing enterotoxin, from healthy carrier examples by PCR.
2. Methods
In this descriptive cross-sectional study, over a period of 5 months, 200 samples from patients admitted to different health care centers of Zabol University of Medical Sciences in 1395 were collected. Then, the samples were inoculated into plastic micro tibial containers (containing BHI medium with 10% glycerol) and transferred to the microbiological laboratory of Zabol Medical College for more precise and diagnostic tests. The obtained samples were cultured linearly on a blood agar base, which was enriched with 5% fresh sheep blood, and incubated for 24 hours at 37°C. Catalase test was used to subtract staphylococci from streptococci and to detect staphylococci from the micrococcus, an oxidation and reduction test was used. Finally, 60 isolates of S. aureus were obtained from a total of 60 collected samples after differential tests (17).
After preparing a 0.5 McFarland solution and cultivating on the Muller Hinton Agar, the antibiotic cefazolin, doxycycline, erythromycin, rifampin, ciprofloxacin, and cotrimoxazole discs with a thickness of 5 mm were placed on the medium with sterile pins and incubated for 24 hours. The diameter of non-growth holes was evaluated using the latest CLSI version.
The following steps were performed to extract genomic DNA:
First, clinical isolates, which were stored with biochemical tests at -20°C, were confirmed. Then, subcultures were performed on a medium of blood agar base with 5% sheep blood on the isolates and finally these were incubated for 24 hours at 37°C. Next, a colony of each cultured isolate, which was divided and numbered according to the number of isolates, was inoculated into 5 mL of bertani. A total of 1.5 mL of the resulting culture medium was poured into plastic microtips with 1.5 plastic and the extraction steps were performed as follows:
These changes included doubling of the centrifuge time of the lysozyme enzyme (2 mg) and adding the final step, including the centrifugal of DNA by ethanol. The DNA was dissolved in 100 µL TE buffer for use in PCR and maintained under -20°C until use.
The primer sequences for F: 5’ F: TTT TTG GCA CAT GAT TTA ATT T 3’ and R: 5’ CAA CCG TTT TAT TGT CGT TG 3’ for amplifying a 541 bp of Ent C gene was designed from the published GenBank sequences with Alleleid 6 Primer Analysis Software and the primer sequences for F: 5’ TGT ATG TAT GGA GGT GTA AC 3’ and R: 5’ TTC GGG AAA ATC ACC CTA A 3’ for amplifying a 306 bp of Ent E gene was designed from the published GenBank.
2.1. PCR Reaction
The reaction was performed in a volume of 25 µL. In this reaction, 1 µL of the template DNA, 0.5 µL of the Taq DNA (5 µL), 0.5 µL of each primer (20 µmol), 2 µL of the Deoxynucleotide triphosphates (dNTPs (2.5 µL) Molar), 2.5 µL of PCR buffer (10 X), and 1.5 µL of MgCl2 (50 mM) were mixed. The final mixture was distilled with 25 µL of water. The reaction was performed in 32 cycles. To investigate the reaction product, 5 µL of electrolyte was transferred to 1% agarose gel. Then, the sample was stained with Ethidium bromide and evaluated.
| Degree | Time | Cycle |
|---|---|---|
| 94 | 5 min | 35 |
| 94 | 10 s | |
| 50 - 60 | 1 min | |
| 72 | 1 min | |
| 72 | 10 s |
Program for PCR Reaction
3. Results
Among 200 specimens, only 60 specimens were identified as Staphylococcus aureus.
The results of this study showed that isolated S. aureus was resistant to antibiotics including cefazolin (7.3%), doxycycline (12.10%), erythromycin (22.3%), rifampin (9.7%), ciprofloxacin (18.10%), and Co-trimoxazole (14.5%).
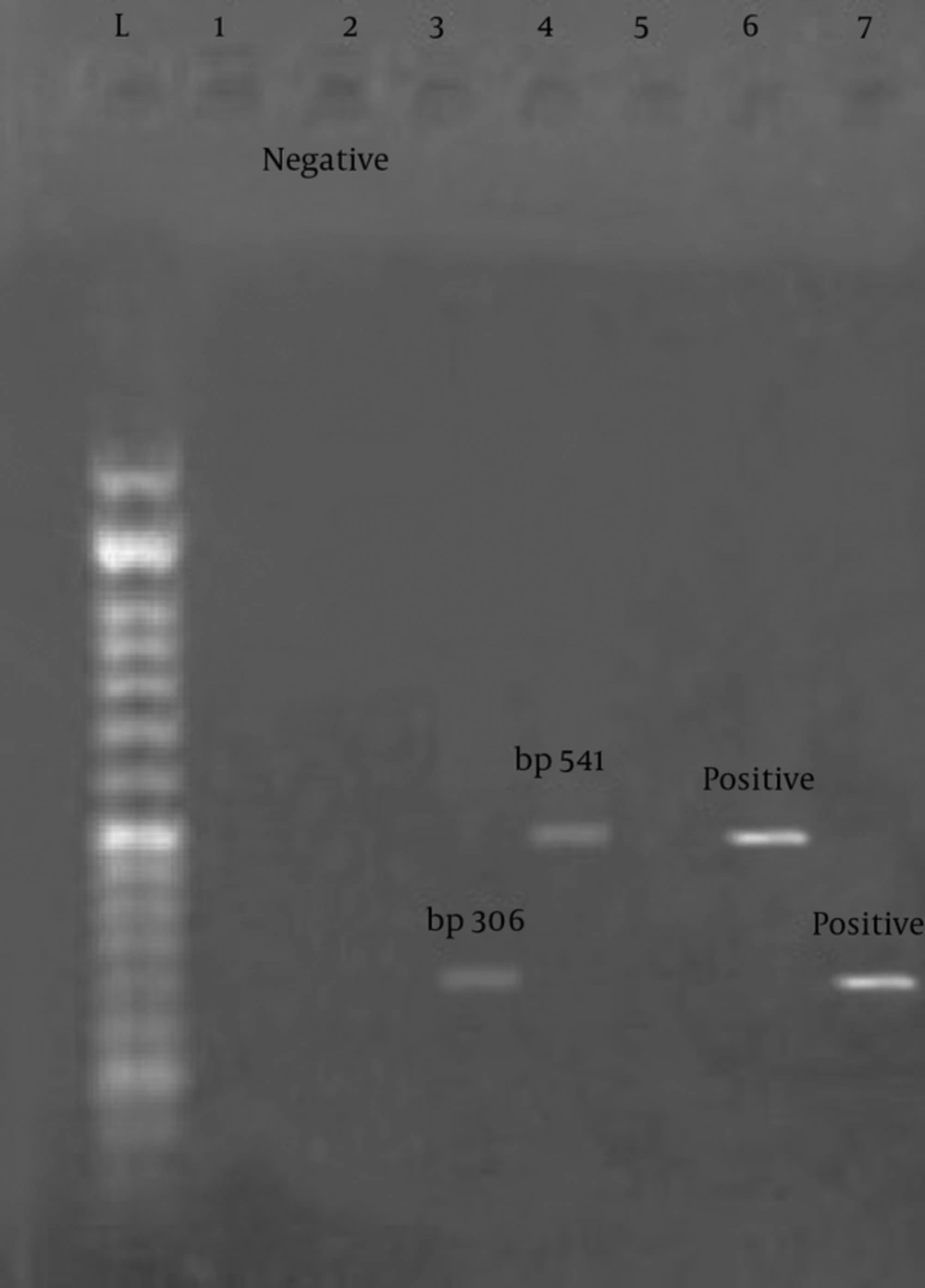
The results of this study showed that among 60 samples tested, 1.5% for the enterotoxin E gene and 4.5% for the enterotoxin C gene were positive. These results indicate the low incidence of enterotoxin E gene in Staphylococcus aureus. Figure 1 shows positive examples of enterotoxin.
4. Discussion
Staphylococcus aureus is found on mucous membranes and skin of mammals, different foods, and surroundings environments, and is the cause of pneumonia through viral infections, bovine mastitis, phlebitis, meningitis, urinary tract infection, local inflammation of the bones, endocarditis, superficial lesions skin, and so on (3).
Staphylococcus aureus produces a wide range of exotoxins, the leading cause of illness in the hosts. Some strains secrete a family of pyrogenic toxins, including enterotoxins, toxins, toxic shock syndrome, and oxalic acid (18).
There is very little information about enterotoxin genes in Iran, and most of the previous research is focused on investigating the antibiotic resistance of this bacterium.
S. aureus secretes 2 types of toxins with superantigen activity, enterotoxins, of which there are 6 antigenic types (named SE-A, B, C, D, E and G) (8).
Therefore, it can be said that enterotoxin genes can also be horizontally transmitted between strains of Staphylococcus aureus. In addition, it is likely that these genetic elements play an important role in the evolution of Staphylococcus aureus as a pathogen. Today SEs are identified and determined by phenotypic techniques including latex agglutination, ELISA, immunomodulation and immuno-diffusion, or genotype techniques including PCR, Multiplex PCR, and Real-time PCR at the level of protein or gene detection.
Ataee et al., investigated the presence of enterotoxin D in patients with rheumatoid arthritis, and the results showed that 60 (55%) and 45 (37.5%) of the samples were positive in synovial and blood samples (19).
The results of the study by Ataee on the enterotoxin-C gene in patients with rheumatoid arthritis in Tehran showed that 66% (33) carry the enterotoxin-C gene (20).
In the study of Noorbakhsh et al., Toxin A, B, and C in synovial samples of patients with rheumatoid arthritis were explored, and the results showed that enterotoxin B, A, and C levels were 18% (n = 10), 39% (n = 22), and 39% (n = 21) that have been reported positive (21).
The results of this study showed that 50% of isolates obtained from Staphylococcus aureus contained enterotoxin genes. The most abundant gene was Sea (30%) followed by Sed (10%), See (8.3%), and Sec (1.6%;) respectively (22).
Saadati et al., identified the secretion of Seas and Seq Sep has of Staphylococcus aureus from healthy carriers, and the results showed that out of 95 healthy individuals carrying the bacterium in their nose, 41 strains (43.1%) were considered as Sea, Sec, or Seq that were positive. A total of 24 strains (25.3%) belonged to the Sea gene, 9 strains (9.5%) were Sec and 8 strains (8.4%) contained Seq gene and 54 strains (56.8%) belonged to other types of bacteria (23).
The results of this study showed that isolated Staphylococcus aureus was resistant to antibiotics including cefazolin (7.3%), doxycycline (12.10%), erythromycin (22.3%), rifampin (9.7%), ciprofloxacin (18.10%), and Co-trimoxazole (14.5%).
In a recent study, Norouzi et al. (2012) showed that out of 80 strains of Staphylococcus aureus, 53 strains (66.25%) contained 1 or more TSST-1 enterotoxin genes. Among the positive samples, 17 strains (32.07%) contained sea gene, 39 strains (73.58%) contained Seb gene, 30 strains (56.6%) had Sec gene, 2 strains (3.77%) containing Sed gene, and 21 strains (39.66 %) contained the See gene (24).
In the study of Zahiri Yegane et al., which the presence of enterotoxin E in 83 patients with rheumatoid arthritis using ELISA and PCR was investigated, the results showed that 13.25% in the PCR method and 40.96 in the ELISA method were positive for enterotoxin E (25).
In the study of Noorbakhsh et al., who examined toxin A, B, and C in synovial samples of patients with rheumatoid arthritis, the results showed that the levels of enterotoxin A, B, and C were reported to be 18% (n = 10), 39% (n = 22), and 39% (n = 21), respectively (26).
In another study by Ataee et al., which identified the enterotoxin C synovial fluid in patients with rheumatoid arthritis, the results of PCR showed that the enterotoxin C gene was observed in 66% (33 cases), while ELISA results reported enterotoxin C level 46% (27).
In the Bokaeian et al., study it was shown that an antibiotic resistance pattern of Staphylococcus aureus that staphylococci were resistant to cefoxitin antibiotics (54.17%), vancomycin (0%), oxacillin (59.41%), cefazolin (25.2%), erythromycin (67.64%), and penicillin (100%) (28).
In a study by Vaez and colleagues, who examined antibiotic resistance patterns of Staphylococcus aureus in Gorgan, the results showed that 104 strains (85.9%) were resistant to methicillin from 121 strains examined. The highest resistance of MrsA strains to penicillin was in 104 strains (100%), Amoxiclav 102 strains (97.6%), cefotaxime 74 strains (71.4%), and erythromycin 67 strains (64.3%), respectively (29).
Parhizgari and colleagues examined antibiotic resistance patterns in Staphylococcus aureus in Ahwaz hospitals. The results showed that out of 255 suspected cases, 180 strains of Staphylococcus aureus were confirmed, in which 59 strains (37.2%) were resistant to methicillin. Resistance to strains of methicillin-resistant Staphylococcus aureus, including chloramphenicol 3.38%, rifampin 45.76%, norfloxacin 89.83%, gentamicin 89.83%, ciprofloxacin 91.52%, azithromycin 88.13%, and co-trimoxazole 86.44% (30).
Musa’i and colleagues examined antibiotic susceptibility of Staphylococcus aureus in Qom hospitals. The results of this study showed the resistance to antibiotics Ciprofloxacin (30.23%), Ofloxacin (27.9%), Norfloxacin (27.9%), levofloxacin (25.58%), Erythromycin (27.9%), Penicillin (86.05%), Tetracycline 30.29%), cotrimoxazole 6.97%), rifampin (20.93%), cefocytosine (37.2%), and clindamycin (23.25%) (31).
In the study of Moradi et al., who examined antibiotic resistance pattern of Staphylococcus aureus in Bandar Abbas hospital, the results showed that the antibiotic resistance was chloramphenicol (7.70%), erythromycin (36.60%), rifampin (16.30%), clindamycin (42.30%), trimethoprim- Sulfamethoxazole (29.80%), cloxacillin (40.40%), ampicillin 21.20%), amikacin (14.40%), gentamicin (21.20%), and methicillin (40.40%) (32).
Faraj Zadeh Sheikh and colleagues examined the prevalence of enterotoxin A gene and antibiotic resistance pattern in Staphylococcus aureus isolates in Ahwaz city. The results showed that out of 222 isolates of Staphylococcus aureus, 102 (45.94%) isolates had an enterotoxin A gene and these isolates had high resistance to ciprofloxacin-methicillin-gentamicin-erythromycin and clindamycin antibiotics (33).
In a study by Tafaroji et al., who examined the frequency of nasal carriers of Staphylococcus aureus and antimicrobial susceptibility patterns in different parts of Masoumeh hospital in Qom in 1395, the results showed that among 90 people (including 25 males and 65 females), 27 were identified as Staphylococcus aureus carriers. There was no significant difference in the prevalence of staph infection among participants in terms of any demographic variables and 89.7% of the samples were sensitized to vancomycin, 93.1% to mupirocin, 96.6% to imipenem, 89.7% to oxacillin, and 3.4% to methicillin (34).
In a study by Mahdiyoun et al., who examined the pattern of resistance of Staphylococcus aureus in Sari and Tehran hospitals, the results showed that the highest resistance to erythromycin were (85.1%), clindamycin (77.6%), cefazolin (77%), ciprofloxacin (75.9%), rifampin (57.5%), doxycycline (55.2%), and cotrimoxazole (24.7%) (35).
Tabaei et al., examined antibiotic resistance patterns in Staphylococcus aureus strains in Imam Reza hospital of Mashhad. The results of this study showed that the most resistance was related to penicillin antibiotics (68.3%), erythromycin (52.2%), clindamycin (42.6%), gentamicin (24%), sulfamethoxazole (15.9%), and ciprofloxacin (13.4%), respectively (36).
4.1. Conclusion
PCR analysis and identification of Staphylococcus aureus strains with the presence of staphylococcal enterotoxin genes gives a basic and more complete view of the risk analysis, therefore, the frequency of enterotoxin producing isolates indicates the risk of pathogenicity in patients. Staphylococcus aureus is fully capable of acquiring mobile genetic elements that encode specific enterotoxins; this is important in preventative and diagnostic applications.