1. Background
Staphylococcus aureus is one of the most important bacterial species that cause food poisoning throughout the world. The poisoning caused by this bacterium is one of the most widespread types of food poisoning and is among the three first-degree poisonings (1). Different investigations suggest that 1% - 5% of infections caused by food materials are related to the consumption of milk and dairy products and 53% of these infections result from the use of contaminated cheese (2). One factor that plays an influential role in the pathogenicity of this bacterium is its capacity to form biofilms. Biofilm increases the resistance of the bacterium against the prescribed antibiotics and elevates the protective mechanisms of the host. This elevated resistance of the bacterium against the mentioned factors plays a key role in the development of nosocomial infections (3). Infections caused by Staphylococcus aureus are recognized as chronic and treatment-resistant infections. In different studies conducted in body and lab environments, it has been proven that Staphylococcus aureus has the capacity to adhere to and penetrate into the epithelial cells of bovine mammary glands (4). Biofilms are structures comprising collections of bacteria; they are surrounded by a polymeric matrix produced by the bacteria themselves and therefore, they can adhere to different surfaces (5). Adhesion of microbial cells to surfaces and their collection to form multilayered cell clusters (biofilm) is a key stage in the development of infections. In this regard, adhesion is considered as one of the most important pathogenic factors in Staphylococcus aureus (6). Instruments such as artificial organs, urethral catheters, and inside-vessel catheters are frequently used for patients being treated in special care divisions of hospitals. Furthermore, bacteria that adhere to these surfaces and form biofilms are hardly killed by antibiotics (3). Researchers have shown that the first stage in infections caused by Staphylococcus aureus is the adhesion of this bacterium to surfaces such as medical instruments, host tissues, etc. This is attributed to a combination of factors external to the cell (e.g. the ability to adhere and form biofilm) (7). This stage is mediated by PIA (Polysaccharide Intercellular Adhesion) whose production is affected by intercellular adhesion proteins such as IcaA, IcaB, IcaC, and IcaD (8, 9). icaA and icaD genes (which include chromosomal genes) have a more significant role in forming biofilm in Staphylococcus aureus and Staphylococcus epidermidis (10). Genes such as fnbA, fnbB (fibronectin binding protein), and clfB (clumping factor) (which includes chromosomal genes) also play a considerable role in elevating the possibility of bacterial adhesion to surfaces (11). Formation of biofilms has significant health-related and economic consequences. It is estimated that 65 percent of nosocomial infections in the US are related to the formation of biofilms and the economic loss caused by biofilms equals more than one billion dollars per year (12). Research shows that adhesion to fibronectin through FnBPs is essential for attacking eukaryote cells (13). In this regard, the significant role of adhesion factors has been investigated in a wide range of diseases caused by Staphylococcus aureus. It has been proven that fnbA and fnbB are significantly influential in the formation of tissues in various pathologic conditions such as eye keratitis (14), osteomyelitis and septic arthritis (15) and substitution in the surface of medical instruments (16) in which Staphylococcus is seen as being an influential disease-producing germ in infections related to implant (injection of medicine or implanting artificial organs) (17). The aim of the current study was to investigate the biofilm-producing genes (icaA, icaD, fnbA, and clfB) in Staphylococcus aureus isolated from local cheese in Maragheh city.
2. Methods
2.1. Samples
In this research, 20 strains of Staphylococcus aureus bacterium extracted from local cheese (soft salty cheese) in Maragheh (already identified through biochemical experiments) were used as samples.
2.2. Extraction of DNA
To extract DNA, a rapid genomic extraction kit (MBST Company, Iran) was used. After extraction of DNA, the samples were read for determination of DNA concentration in ng/L using a Nanodrop device and on the basis of priority of reading indices, the ratio of 260/280 nm, the ratio of 260/230 nm, and optical density (OD) were read for performing PCR, respectively.
2.3. PCR Test to Detect Thermonuclease Gene (nuc)
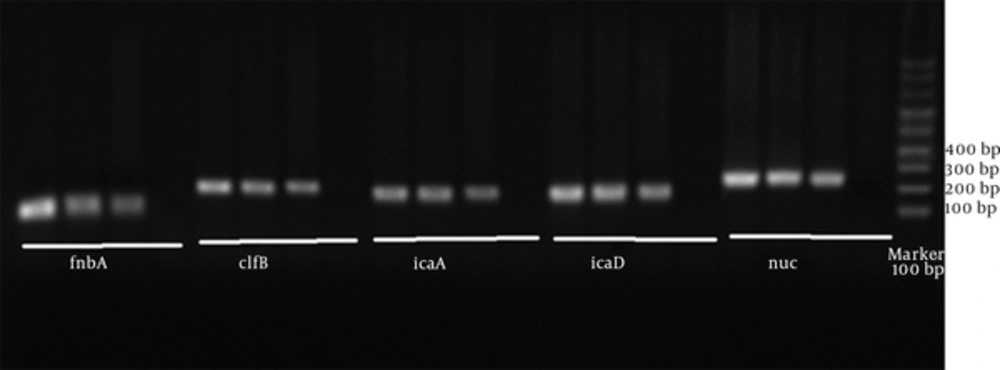
The polymerase chain reaction was performed in 25 µL volume, including extracted DNA containing 1 µL (50 ng) Master mix, 12.5 µL PCR, 0.4 µM specific primers (0.5 µL from each of the forward and reverse primers) (18) (Table 1) and 10.5 µL of double distilled water. PCR conditions are presented in Table 2. PCR product in 1.5% agarose was electrophoresed and imaged using gel document. S. aureus PTCC1112 DNA extract was used as a positive control. Double distilled water was used for negative control. S. aureus isolates were considered as S. aureus after a definite diagnosis of Thermonuclease gene with a length of approximately 279 bp.
| Gene | Primer Sequence | PCR Product, bp |
|---|---|---|
| nuc | 5’-GCGATTGATGGTGATACGGTT-3’ | 279 |
| 5’-AGCCAAGCCTTGACGAACTAAAGC-3’ | ||
| fnbA | 5’-CATAAATTGGGAGCAGCATCA-3’ | 128 |
| 5’-ATCAGCAGCTGAATTCCCATT-3’ | ||
| clfB | 5’-ACATCAGTAATAGTAGGGGCAAC-3’ | 203 |
| 5’-TTCGCACTGTTTGTGTTTGCAC-3’ | ||
| icaD | 5’-ATGGTCAAGCCCAGACAGAG-3’ | 198 |
| 5’-AGTATTTTCAATGTTTAAAGCAA-3’ | ||
| icaA | 5’-ACACTTGCTGGCGCAGTCAA-3’ | 188 |
| 5’-TCTGGAACCAACATCCAACA-3’ |
| Cycle | Step | Temperature, °C | Time, Min |
|---|---|---|---|
| 1 | primary denaturation | 94 | 4 |
| 32 | Denaturation | 94 | 1 |
| Annealing | 55 | 1 | |
| Extension | 72 | 1 | |
| 1 | Final extension | 72 | 10 |
2.4. PCR Test to Identify the Biofilm-Producing Genes Under Investigation
To perform PCR, the Taq DNA polymerase kit (QIAGEN Company, Catalogue no. 201223) was utilized. In a 25 µL volume, this reaction includes 12.5 µL of double distilled water, 2.5 µL of buffer, 1 µL of dNTPs, 3 µL of primers (1.5 µL of forward primer and 1.5 µL of reverse primer), 0.75 µL of MgCl2, 0.25 µL of Taq DNA Polymerase, and 5 µL of extracted DNA. The sequence of specific primers used for icaA, icaD, clfB, and fnbA genes (19-21) is indicated in Table 1. The proliferation of target genes in thermocycler was carried out through the following timetable (Table 3).
| Cycle | Step | Temperature, °C | Time |
|---|---|---|---|
| 1 | Primary denaturation | 94 | 4 min |
| 35 | Denaturation | 72 | 30 sec |
| Annealing | 55 | 30 sec | |
| Extension | 72 | 1 min | |
| 1 | Final extension | 72 | 10 min |
PCR products related to the proliferation of each of the adhesion genes under investigation were studied through electrophoresis on agarose gel, which was observed by gel documentation. In order to show the proliferated pieces, DNA Ladder 100 bp was utilized. Extracted Staphylococcus aureus PTCC 1112 DNA was used as positive control. Double distilled water was used as negative control.
3. Results
17 strains (85%) out of 20 bacterial strains were recognized as positive species-specific Staphylococcus aureus in terms of thermonuclease (nus). The frequencies of each of the genes of the investigated strains were icaA (76.47%), clfB (76.47%), icaD (58.82%), and fnbA (64.7%) (Figure 1). 5 strains (29.41%) simultaneously included all the genes under investigation (Table 2).
| Gene | Number of Isolates | Combination of Genes | Number of Isolates |
|---|---|---|---|
| icaD | 10 | icaA + clfB | 1 |
| icaA | 13 | icaA + fnbA | 1 |
| clfB | 13 | icaA + icaD + clfB | 7 |
| fnbA | 11 | icaA + icaD + fnbA | 1 |
| icaA + clfB + fnbA | 2 | ||
| icaD + clfB + fnbA | 1 | ||
| icaA + icaD + clfB + fnbA | 5 | ||
| Total | 17 |
4. Discussion
This research reported the frequencies of icaA (76.47%), clfB (76.47%), icaD (58.82%), and fnbA (64.7%) genes. 5 strains (29.41%) simultaneously included all the genes under investigation. Cifter et al. (2009) reported that 25.42% of Staphylococcus aureus bacteria extracted from mastitis included simultaneously icaA and icaD genes, 27.11% of them included icaA gene, and 64.4% included icaD gene (21). The frequency of icaD gene is in line with the findings of the present study. Shojaei et al. (2014) reported that the frequencies of icaA, icaD, clfB, and fnbA in Staphylococcus aureus extracted from raw milk were 59.18%, 38.77%, 32.6%, and 69.38%, respectively (11). This contrasts with the findings of the present study. Basanisi et al. (2016) found that 94.6% of Staphylococcus aureus bacteria extracted from dairy products of sheep and goat in the south of Italy included icaA gene (22). The widespread distribution of Staphylococcus aureus strains including biofilm genes in milk increases the risk of the presence of these strains in food products. Investigating the capacity of adhesion and formation of biofilm as well as the genes playing a role in the formation of biofilms in different Staphylococcus aureus strains can provide invaluable information for better understanding of the complex process of biofilm formation and infections resulting from these organisms (19). This bacterium has a high ability to form biofilms, which can be of significance in the development of chronic infections and the development of strains resistant to antibiotics. Furthermore, through different mechanisms of transferring resistance against medicines, bacteria found in biofilms can elevate the resistance of infection-producing bacteria in special care divisions of hospitals; this phenomenon can be considered as a serious threat for patients being treated in medical centers. When compared to the findings of other studies, the results of the current research show a difference in the distribution of the genes that play a role in the formation of biofilms among the strains of Staphylococcus aureus; this could be due to the geographical differences and the differences in ecological origins of the extracted strains (Milk, humans, and other animals).
According to the findings of the current research and other similar investigations, none of the samples of the Staphylococcus aureus samples was negative in terms of the presence of the four genes being investigated. This shows the high ability of this bacterium in forming biofilms. It is suggested conducting more research on the precise combination of the structure of the Staphylococcus aureus biofilm so that this structure can be more precisely targeted by the medicines. In addition, protein combinations of the structure of biofilm capsule of this bacterium can be used as a vaccine against biofilm infections.