1. Background
Johne’s disease, or paratuberculosis, is an incurable chronic infectious disease in domestic and wild ruminants worldwide. This disease causes granulomatous enterocolitis, lymphangitis, lymphadenitis, and progressive weight loss. Symptoms of more advanced clinical diseases include ventral edema and 'water hose' diarrhea, but also remain bright and alert with a good appetite (1). Economic losses can be substantial when a beef or dairy herd becomes infected with Mycobacterium avium subsp paratuberculosis (MAP). The most apparent losses are from symptomatic animals that are culled. However, premature culling of positive asymptomatic animals results in the loss of potential income from reproduction show, or sales (2). Disease agent is MAP (3, 4). The main source of infection is infected animal feces. In addition, direct contact of susceptible animals with infected feces is a main factor in prevalence and spread of disease. Except feces, the agent of disease is isolated from different organs of infected animals (5). Hines et al., (1987) showed that MAP is able to spread throughout the body as well as intestine. M.avium complex is one of the most important pathogenic species that consist of three subspecies: M. avium subsp avium, M. avium subsp paratuberculosis, and M. avium subsp silvaticum.
Theses subspecies can be distinguished from each other by IS900 locus (6). Johne’s disease was initially diagnosed in 1960 among the imported cows in Iran, however, due to limited studies, the exact incidence rate and disease frequency have not been indicated yet (7). The highest predisposition of stock morbidity is at the age of below 30 days, however, due to the length of incubation period of disease, clinical signs of disease appear after the age of 2 years (8). Recently, it has been shown that MAP causes crohn’s disease in humans, which leads to progressive weight loss and granulomatous lesion in different organs, especially in the digestive system. There are many methods for isolation of the agent of Johne’s disease, however, none of them are worth independent diagnosis, and a complementary method is required (7). Although fecal culture has been used as a gold standard method for the detection of MAP, due to factors like slow rate of growth of organism in culture medium, time-consuming incubation period, probability of pollution of media, difficulty associated with fecal culture of animals that excreted less than 100 organisms per gram of their feces, and also inefficiency of this method in detecting subclinical and carrier animals, it is limited and the emerging negative outcomes cannot completely reject the presence of Johne’s disease in a herd (9). Laboratories and veterinarians have sought alternative diagnostic tests for MAP due to limitations in the sensitivity of fecal culture as well as the long time required for culture results (2). Due to the fact that carrier and subclinical cows in Johne’s disease do not excrete the organism through feces, blood sampling and performing exact examination are necessary in these animals (10). Excretion of the organism may occur for prolonged periods (1 to 2.5 years) before the onset of clinical disease. Thus, early diagnosis of infected animals is needed to avoid the spread of the disease, due to the fact that control depends on detection and culling of infected animals as early as possible (11). At the moment, ELISA and PCR methods, which possess high sensitivity and specificity in searching agents of disease in suspected and infected cases and also in asymptom and carrier animals, are important (12). PCR has been used to improve the identification of microorganisms, especially when traditional microbiological detection methods have serious limitations (11). IS900 is an insertion sequence of 1,451-bp characteristic of M. avium subsp. paratuberculosis that encodes for a protein of 399 amino acids and repeats at 15 to 20 copies per genome (13). IS900 has been demonstrated to be a useful target for PCR amplification to aid in the diagnosis of M. avium subsp. paratuberculosis in biological samples (14). Although IS900-like sequences are found in other bacteria, the stringent selection of IS900-like specific primers avoids the amplification of IS900-like elements (15, 16).
The aim of this study was to compare bovine fecal and buffy coat samples for diagnosis of Johne’s disease based on PCR and finding out which one was more suitable for detection of MAP.
2. Methods
Feces and blood samplings were performed from 100 suspected cows with clinical signs of Johne’s disease such as weight loss and diarrhea from dairy herds in North West of Iran, Tabriz from March 15, 2015 to February 15, 2016. From each cow, a blood sample (using heparine venoject tube) and a feces sample were taken individually. Then, samples were transferred to a laboratory icy environment.
DNA was extracted from both fecal and blood samples by CTAB and proteinase K method (17). Designed specific primers IS900 (MWG company) were used for PCR test (Forward F90: 5´-GTTCGGGGCCGTCGCTTAGG-3´ and Reverse R91: 5´-GAGGTCGATCGCCCACGTGA-3´) (18). Then, PCR product was electrophoresed in gel agarose and presence of MAP was confirmed by showing PCR expected band. Standard bacterial strain ATCC 19698 was used as a positive control in PCR test (18). Amplification was undertaken in a programmable thermocycler under the following conditions: 1 initial cycle of denaturation at 94°C for 5 min; 40 cycles of denaturation at 94°C for 1 min, annealing at 65°C for 30 s, and extension at 72°C for 1 min; and a final extension at 72°C for 10 min. PCR product in 1.5% agarose was electrophoresed and imaged using gel document. Agreement among tests was measured by calculating the kappa coefficient (19). To compare the values of a categorical variable when these values are obtained from two methods of measurement i.e. assessing of agreement between two different methods, we can calculate Cohen Kappa’s coefficient. Based on this coefficient, we can decide in agreement level between two methods.
3. Results
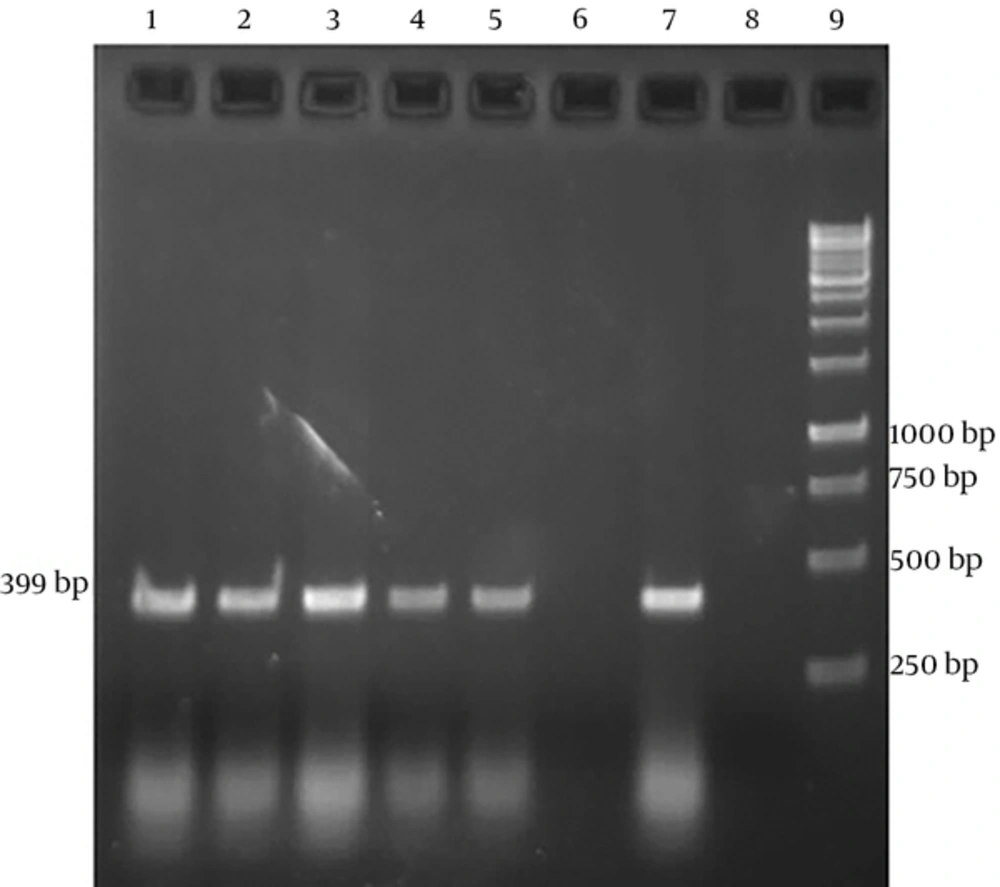
Sampling was performed among 100 suspected cows with clinical signs of Johne’s disease such as weight loss and diarrhea. In this study 4 and 9 samples were positive in PCR test from buffy coat and feces, respectively (Table 1). Positive PCR products using specific IS900 primers were detected as a 399bp product (Figure 1). After calculating, Cohen’s kappa coefficient was 0.593 (CI %95=0.245 to 0.941). Therefore, the agreement level between buffy coat and fecal PCRs is moderate.
| Sample | Buffy Coat-PCR | Fecal-PCR |
|---|---|---|
| Positive | 4 | 9 |
| Negative | 96 | 91 |
| Total | 100 | 100 |
Mycobacterium avium subsp paratuberculosis specific amplicons (399bp) by PCR using IS900 specific primers. Lanes 1-3 show fecal PCR products and lanes 4 and 5 show buffy coat PCR products. These lanes create bands within 399bp, which correspond with positive control (Lane 7). Lane 6 shows negative result. Lane 8 indicates negative control (no DNA). Lane 9 is DNA ladder (1 kb).
4. Discussion
Johne’s disease is economically important as an incurable chronic infectious disease in ruminants worldwide. Clinical signs of disease in cows do not appear before the age of 2 years, and it usually occurs within the age of 2-6 years, therefore, detection of carrier and disease animals with suitable and sensitive diagnostic methods is important from an epidemiological point of view (8). Lack of acceptable laboratory methods to detect subclinical infected animals is a major problem in controlling Johne’s disease and all commonly used bacteriological and immunological methods have a high error rate for the detection of this disease. No single method with reliable results exists for diagnosis of disease in all stages and in these methods there are always false positive and negative results (10). MAP is resistant to intracellular degradation and remains viable inside macrophages for several weeks and can even grow inside macrophages. The organism can transfer to the liver, spleen, uterus, udder gland, reproductive organs, and other organs through macrophages. In addition, in severe infections, it leads to severe bacteremia and infection of phagocyte cells. The organisms are phagocyted by macrophages and then a lot of them multiply into macrophages and penetrate into submocusal layer of intestine. This leads to reduction of absorption, chronic diarrhea, and malabsorption. Thus, the excreted organism results in spread of disease through feces as a major source of infection (5). The organism is isolated from feces, lymph nodes, udder gland, uterus, reproductive system, blood cells, milk, and some organs such as kidney (4). In the current study, 4% and 9% of samples were positive in PCR test from buffy coat and feces, respectively. Taylor et al. (20), isolated MAP from milk. Larsen et al. (21), isolated the organism from reproductive system and sperm. Hines et al. (22), isolated disease agent from cow serum. Sambrook et al. (23), isolated the organism from buffy coat of cows. Hurley et al. (24), succeeded to detect the organism from feces of cow by molecular method. Whipple et al. (18), isolated MAP from cows suspected of Johne’s disease by PCR method. Bull et al. and Ellingson et al. (6, 25), isolated the organism from infected animals by PCR method. Although the disease has spread all over the world, it seems to be growing in some countries. Disease occurrence is growing in areas with a temperate climate and rarely in tropical and subtropical countries, especially in Iran. The incidence rate of disease is high in animals that are kept in compact condition. In the present study, more positive cases were among compact herds. One of the major reasons for the more frequent occurrence of Johne’s disease in dairy cows compared with beefs is that beefs are bred in larger areas; therefore, they are less exposed to sources pollution such as feces. In the field situation, disease agent primarily transfers through contaminated food and water by feces of infected animals that excrete the organism (10).
The major problem in detecting Johne’s disease is accurate diagnosis of resistant animals, which have never been infected, however, their cutaneous and serological tests are positive and in some cases animals that are in the intermediate stage of disease and excrete the organism through feces may show negative results in serological tests. MAP can be detected from feces and body fluids by PCR method. Regarding the fact that fecal culture is a time consuming method and it cannot identify subclinical and carrier animals, and since it needs more speed in control, prevention, and eradication of disease, PCR method is very important. In our study, the little number of positive cases in associated cases with blood (buffy coat) can result from many factors such as presence of inhibitors in blood and low bacterial concentration in it. According to the findings of the present study, it can be concluded that PCR is a rapid, sensitive and accurate method with both high sensitivity and specificity in diagnosis of asymptom carrier and subclinical infected animals. Therefore, PCR method plays a major role in prevention and control of Johne’s disease. According to kappa coefficient, agreement between two tests based on calculated amount showed moderate agreement. Thus, replacement of buffy coat PCR with fecal PCR was not recommended. Most of the previous studies have recorded high agreement between fecal PCR and fecal culture (as a gold standard test) (2), therefore, using fecal samples based on PCR for diagnosis of Johne’s disease is more efficient.