1. Background
Breast cancer (BC) is the most prevalent type of malignancy in females, with two patterns, including hereditary and sporadic (1). The environmental risk factors, consist of ionizing radiation (IR), carcinogenic components, heterocyclic aromatic amines, alcohol consumption, and free radicals (2, 3). Genetic factors are compound conditions of BC (4). During DNA damage, eukaryotic cells use appropriate processes to repair multiple forms of DNA damages to protect genome stability and integrity. Repairing pathways have significant roles in maintaining genomic integrity in comparison with carcinogenic components and mutagens (5). During exposure of DNA to IR, the double-stranded DNA may be broken and this increases the risk of BC progress (6). In humans, base excision repair (BER) and Nucleotide Excision Repair (NER) comprised of the two most common DNA repair mechanisms (7, 8). In BER systems, there are eleven DNA damage special proteins that contribute to a specific function. Among these components, X-ray repair cross-complementing group 1 (XRCC1) participates in the BER system (9, 10). Another significant molecule, excision repair cross-complementation 1 (ERCC1), which encodes an ERCC1 protein, is involved in the NER pathway, eliminating significant DNA damage generated through environmental agents including toxic compounds or ultraviolet (10). Genetic variations, such as single nucleotide polymorphism (SNP) may exchange amino acids or modify DNA conformations, which is related to cancer risk or human disorders (11, 12). Single nucleotide polymorphism molecular analysis could facilitate prognosis, diagnosis, and remedy of human disorders (13). With the crucial roles of DNA repairing machinery components in human genome integrity, it is possible to reveal significant appearances of nucleotide changes in the DNA repair genes and the risk of malignancies (14). Preliminary investigations have shown several SNPs in the XRCC1 and ERCC1 genes involved in tumorigenesis. According to previous considerations, findings have revealed contradictory results regarding several diseases and different ethnic geographical regions (15-17). To provide greater insight regarding the association between particular SNPs in the XRCC1 and ERCC1 genes in the tumorigenesis of BC, this study aimed at evaluating the associated between XRCC1 (rs25487) and ERCC1 (rs3212964) gene polymorphisms and the risk of sporadic BC in females of the northwestern of Iran.
2. Methods
2.1. Patients and Controls
In a case-control investigation, we recruited 100 females, who had been referred to the Omid hospital of Urmia, Iran. All cases were diagnosed as having histologically sporadic BC (mean age 44.69 ± 5.12 years), and 100 age-matched healthy females (mean age 44.5 ± 6.5 years), who attended the related hospital for conventional physical assessment, were registered as the control group, during June 2015 to April 2017. All individuals were from the same geographic province. The clinical features of each participant, including history of smoking, drinking, BRCA1/BRCA2 gene mutation status, and other malignancy history, was collected through a survey and signed informed consents were obtained from all participants. Exclusion criteria comprised of having metastasized carcinoma or identified mutations in the BRCA1/BRCA2 genes, and having received a radiotherapy or chemotherapy regimen. This study excluded all samples, which did not have the criteria of the community before genetic variant analysis. The ethical review board of Omid hospital of Urmia approved the investigation.
2.2. Genetic Variant Analysis
We selected two common SNPs in XRCC1 (rs25487) and ERCC1 (rs3212964) genes to evaluate the genetic susceptibility of sporadic BC. The genomic DNA was collected from white blood cells using the salting-out method reported by Miller et al. (18). For quantitive and qualitative appraisal of extracted DNA, NanoDrop I (Thermo Scientific Fisher, USA) and 1% agarose gel electrophoresis were used, respectively.
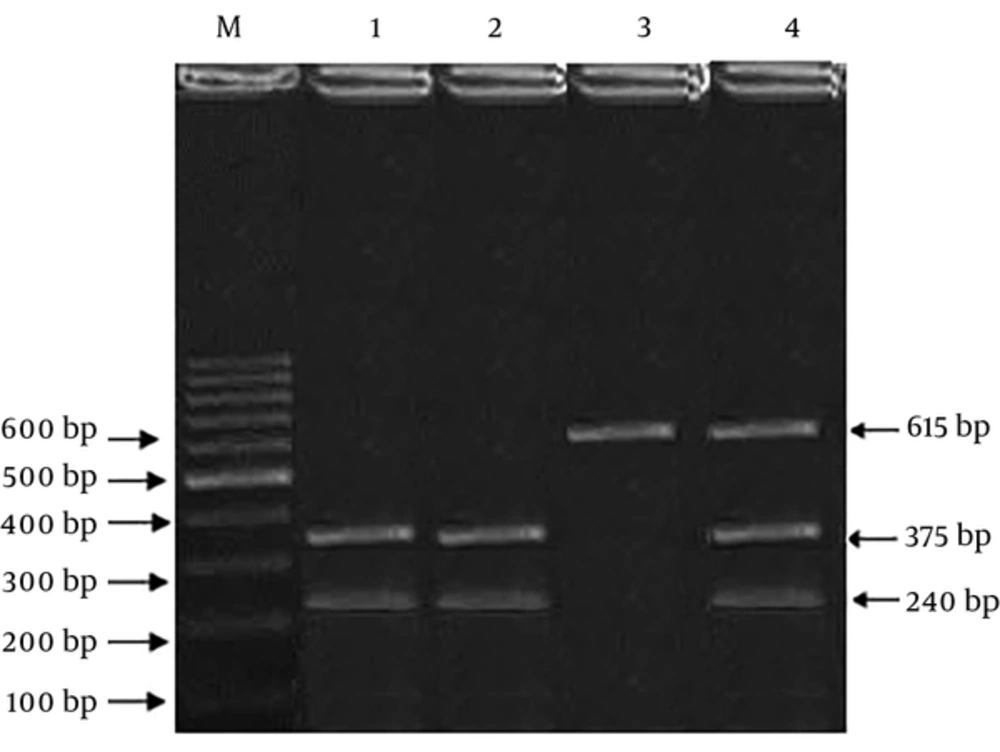
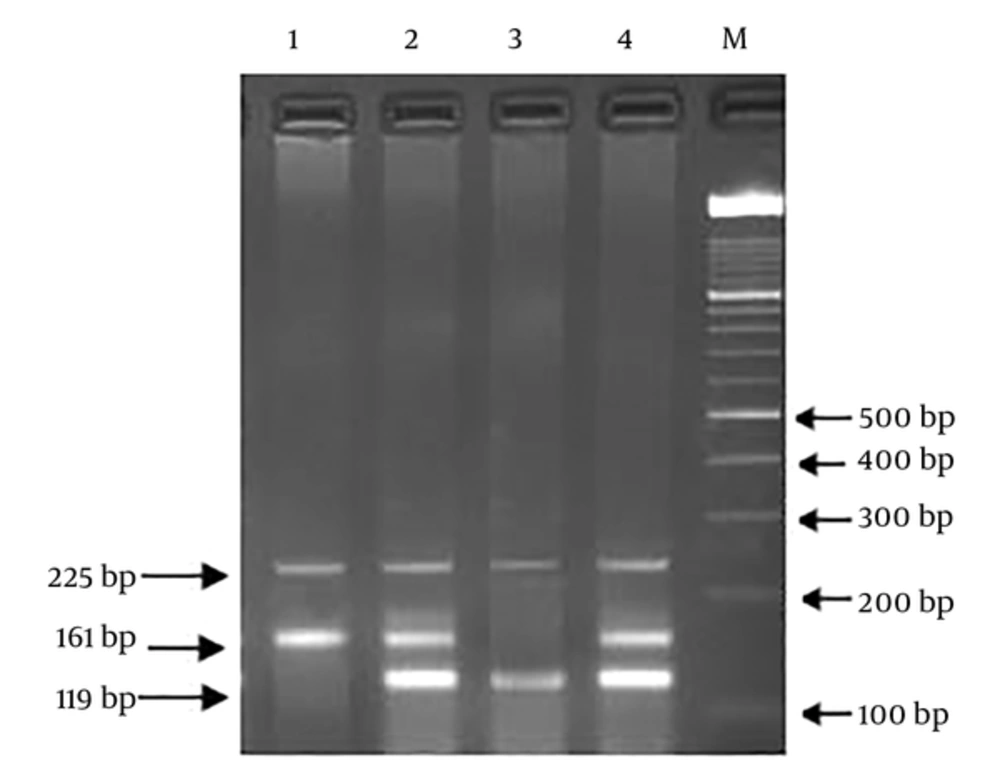
For determining the genotype frequencies, we used Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP) and ARMS-PCR (Amplification-refractory mutation system) techniques. The primer sequence of rs25487 was (F): 5’-TTGTGCTTTCTCTGTGTCCA-3’, (R) 5’-TCCTCCAGCCTTTTCTGATA-3’ and digested PCR products were obtained using Restriction Enzyme MSP I (Thermo Scientific Fisher, USA), according to the company protocols and separated on 2% agarose gel. For the amplification of the rs3212964 region, Tetra primer sequences were used, including; (Forward Inner): 5’-TCACATCCTCTCTCCCGTAGGGATCA-3’, (Reverse Inner):5’-GGGAAAGAGGGCTTGAGGAATTATAAGTC-3’, (Forward Outer): 5’-GTGACCTCCAACCTCTACCCAGTTCTC-3’, (Reverse Outer): 5’-ACATTAGAGCTGAGACCCAAAGGAGGAT-3’. The polymorphic region extended through PCR Thermal Cycler (Eppendorf, Germany) in a 25-µL final volume, comprised of 50 ng of genomic DNA, 12.5 µL of 2X Master Mix Red (Ampliqon, Denmark), and each of the primers at a final concentration of 10 pmole. Annealing temperatures were as follows: rs25487 at 58°C and rs3212964 at 60°C. During PCR products digestion, rs25487 (G: 375 + 240 bp, A: 615 bp) and rs3212964 (A allele: 161 bp, G allele: 119 bp, two outer primers: 225 bp (gene control) fragments were generated. To confirm genotype frequencies, 15% of specimens were chosen for double assessment. Additionally, 10% of random individuals with the two SNPs were verified by direct sequencing.
2.3. Statistical Analysis
Relationships between the two SNPs and sporadic breast cancer risk was evaluated by chi-square test, odds ratios (ORs) and their 95% confidence intervals (CIs), which were calculated using the logistic regression model. Statistical analysis was performed using the SPSS software version 22.0 (SPSS Inc., Chicago, IL, USA). P values of < 0.05 were considered as statistically significant.
3. Results
The genotype frequencies of the two SNPs are shown in Table 1. The current findings did not reveal statistically significant differences between case and control groups (P = 0.364). In order to assess whether an association was present between gene polymorphism and the risk of sporadic BC in codominant, dominant, and recessive genetic models, the genotype frequencies of each group was consolidated in new heredity models, then, odds ratios and 95% CI were calculated. The AG/AA+GG genotypes did not demonstrate a risk of sporadic BC (P = 0.254; OR:1.385; CI:0.791 - 2.425; Table 1).
In addition, AA/AG+GG and GG/AA+AG genotype models of the XRCC1 gene did not show a significantly increased risk of sporadic BC (P > 0.05; Table 1).
Of the 100 patients studied for the XRCC1 gene polymorphism, the frequency of the G mutant allele did not show a statistically significant difference between the two groups (P = 0.202; OR: 1.312; CI: 0864 - 1.994; Table 1).
| Gene Polymorphism | Case Group (n = 100)a | Control Group (n = 100)a | Total (n = 200) | OR | CI 95% | P Value | |
|---|---|---|---|---|---|---|---|
| Up | Down | ||||||
| XRCC1 (rs25487; A>G) | |||||||
| Codominant | 1.385 | 2.425 | 0.791 | 0.254 | |||
| AG | 48 (55) | 40 (45) | 88 | ||||
| AA+GG | 52 (46) | 60 (54) | 112 | ||||
| Dominant | 1.500 | 2.626 | 0.857 | 0.155 | |||
| AA | 40 (44) | 50 (56) | 90 | ||||
| AG+GG | 60 (55) | 50 (45) | 110 | ||||
| Recessive | 1.227 | 2.986 | 0.504 | 0.651 | |||
| GG | 12 (55) | 10 (45) | 22 | ||||
| AG+AA | 88 (49) | 90 (51) | 178 | ||||
| Frequency | 1.312 | 1.994 | 0.864 | 0.202 | |||
| Frequency of G allele | 72 (55) | 60 (45) | 132 | ||||
| Frequency of A allele | 128 (48) | 140 (52) | 268 | ||||
| ERCC1 (rs3212964; A > G) | |||||||
| Codominant | 1.042 | 1.825 | 0.595 | 0.886 | |||
| AG | 43 (51) | 42 (49) | 85 | ||||
| AA+GG | 57 (50) | 58 (50) | 115 | ||||
| Dominant | 0.814 | 1.429 | 0.463 | 0.473 | |||
| AA | 39 (47) | 44 (53) | 83 | ||||
| AG+GG | 61 (52) | 56 (48) | 117 | ||||
| Recessive | 1.348 | 2.887 | 0.630 | 0.440 | |||
| GG | 18 (56) | 14 (44) | 32 | ||||
| AG+AA | 82 (49) | 86 (51) | 168 | ||||
| Frequency | 1.213 | 1.820 | 0.808 | 0.352 | |||
| Frequency of G allele | 79 (53) | 70 (47) | 149 | ||||
| Frequency of A allele | 121 (48) | 130 (52) | 251 | ||||
aValues are presented as No. (%).
In the current investigation, a statistically significant difference was not shown in genotype frequencies of the ERCC1 gene between sporadic BC patients compared to controls (P = 0.666). Similar to the XRCC1 gene polymorphism analysis, the genotype frequencies of the ERCC1 gene were assessed for three heredity models. The AG/AA+GG genotypes did not increase the risk of sporadic BC (P = 0.886; OR: 1.042; CI: 0.595 - 1.825; Table 1).
In addition, AA/AG+GG and GG/AA+AG heredity models of ERCC1 gene did not show a significantly increased risk of sporadic BC (P > 0.05), Table 1. Also, the frequency of the G mutant allele was higher in the case group (n = 79) compared to the controls (n = 70), while the difference between groups was not statistically significant (P = 0.352; OR: 1.213; CI: 0.808-1.820; Table 1).
4. Discussion
The current investigation studied the association of two SNPs in the XRCC1 and ERCC1 genes with the risk of sporadic BC in females of northwestern Iran.
The findings revealed that the XRCC1 (rs25487) and ERCC1 (rs3212964) gene polymorphisms were not associated with an enhanced chance of sporadic BC. The present investigation is the beginning of a case-control survey to evaluate the association between two nucleotide changes in the BER pathway related-genes and the risk of sporadic BC in Iranian females.
During the DNA base pair repair procedure, the XRCC1 gene was implicated in the change of DNA damages and single-strand DNA disputes through DNA ligase III enzyme, at its carboxyl and DNA polymerase beta and poly ADP-Ribose polymerase at the site of the damaged DNA (15, 19). The XRCC1 function is needed for DNA repair and genome integrity (20). DNA repairing deficiency may lead to accumulated DNA damages and mutations, which thereafter beginning conditions such as malignancies (21). The XRCC1 gene polymorphism is found inside the XRCC1 BRCA1 carboxyl-terminal domain (BRCT I) and may be modified for protein construction and function. Previous investigations have reported that the genetic variants of the XRCC1 (rs25487) gene are associated with the risk of BC (22, 23). Of note, the documents are not compatible with the current investigation of Iranian females in the Western Azerbaijan province. Zhu et al. proposed that the XRCC1 (rs25487) gene polymorphism may increase the risk of BC (15). In addition, Luo et al. revealed that the G allele in XRCC1 increased the risk of BC and may contribute to tumorigenesis in Chinese females (24). Although epidemiological investigations of the XRCC1 gene polymorphism with the risk of BC have been assessed in various populations, the findings are contradictory and ambiguous. Similar to the current findings, Al Mutairiet al.’s study did not reveal a positive association between XRCC1 (rs25487) gene polymorphism with the risk of BC in Saudi females (25). Ding et al.’s study on Chinese females showed an increased risk of XRCC1 (rs25487) gene polymorphism with BC (26), which is in contrast with the current findings, and a meta-analysis published by Wu et al. on 44 case-control investigations (27). Taken together, the current results did not suggest that the XRCC1 gene polymorphism plays a significant role in the initiation of BC.
Similar to the current findings, previous investigations did not show an association between the ERCC1 (rs3212964) gene polymorphism and the risk of BC (15, 28). Furthermore, ASE-1 has been found in an antisense orientation and overlaps with the ERCC1 gene and is likely involved with RNA polymerase I transcription complex. Aiub et al. showed the BER mechanism was implicated in DNA damages repair, as a result of compounds such as N-nitrosodiethylamine (29). However, preliminary studies did not reveal a significant association between nucleotide change in the ERCC1 gene and the risk BC.
The contradictory results may be due to the low demographic power, inadequate sample size, different allele frequencies, differences in the genetic background of the population (ethnic and racial). In addition, another possibility may be social behavior and environmental factors affecting the risk of BC. However, more investigations are needed to evaluate the role of genetic variations of BER mechanism genes in the pathogenesis of BC. The current results provided evidence that XRCC1 (rs25487) and ERCC1 (rs3212964) gene polymorphisms may implement both independent and interactive impacts on the progress of breast cancer. While BC is considered as a heterogeneous disease, large-designed geographic investigations endeavoring to consider gene-gene and gene-environment interactions are needed for the future studies.