1. Background
Staphylococcus spp. belongs to the Micrococcaceae family and is a gram positive, non-spore bacterium. This bacterium is one of the important germs in medicine and veterinary medicine and some members of this genus are among the most common factors causing mastitis in cows and sheep (1). Some members of this genus are on the skin, skin glands, and mucosal members of animals, and through this they can transfer to animal products and environmental resources. Staphylococcus aureus is a species that not only causes pathogenicity in humans and animals, it can also play an important role in food spoiling (2). This bacterium is the most common factor of bacterial food poisoning and usually contaminates foodstuffs during preparation (3). Foodstuffs including milk, cheese, butter, creamy sweats etc. are a suitable media for S. aureus (4). The types of food that cause food poisoning differ from one country to another. For example, cheese was the most common factor of staphylococcal food poisoning between 1999 - 2000 in France (4). According to different studies, 1% - 5% of food borne intoxications are associated with milk consumption and dairy products, 53% of which are due to consumption of contaminated cheese. Cheese production has been practiced since a long time ago in Iran and still, there is much tendency to use local or unpasteurized cheese. A total of 20% of the milk produced in milk industries turn into cheese in Iran, much of which is devoted to the production of local cheese (5). One of the important hygienic problems in the production of local cheese is related to the abundance of microbial load due to the use of raw milk. Raw milk, after milking and during cheese production stages, is contaminated by various microbes such as S. aureus (6). Furthermore, this bacterium can contaminate milk via breast suffering clinical or subclinical staphylococcal mastitis. This bacterium can grow and duplicate in milk and its products during production and storage stages, which leads to the production of pathogenic enterotoxins (7). S. aureus has many virulence factors, the most important of which are enterotoxins and toxic shock syndrome toxin (TSST-1) and have a significant impact on the host (8). Expansion of antibiotic resistance is another problem with which physicians deal, and due to antibiotic resistance strains emersion in S. aureus, the number of antibiotics gradually decreases for the treatment of its infections. Most of S. aureus isolates are resistant to penicillin, some of them are resistant to methicillin and nafcillin, and a few of them are resistant to vancomycin (9).
2. Objectives
Due to the importance of this subject and also high production and consumption of local cheese in Iran, the current study aimed to determine the presence of toxic shock syndrome toxin (TSST-1) and methicillin resistance (mecA) producing genes of S. aureus strains isolated from local cheese in northwest of Iran.
3. Methods
3.1. Samples
The present study was performed in the Microbiology Laboratory at the Islamic Azad University, Maragheh branch, in May 2017. In this study, we isolated 22 S. aureus strains from 100 local cheese in northwest of Iran, which already identified with biochemical experiments (e.g. Baired parker and Mannitol salt agar, Coagulase and Voges proskauer tests and gram staining).
3.2. DNA Extraction
DNA extraction was performed on 22 cultured isolates of S. aureus in brain heart infusion (BHI) (Merck, Germany) medium. One mL of bacterial culture centrifuged in 5000 g for five minutes and supernatant was poured off. Subsequent to an addition of 1 mL lysis buffer (Tris 1 M (ph = 7.5), NaCl 5 M, EDTA 0.5 M and C-TAB 2%) on mixed pellet, it was put into 85°C for 30 minutes (in water bath). In the next step, the supernatant was separated and 0.5 µL RNase was added to it. Later, it was kept at 37°C for 30 minutes. Then, an equal volume of isopropanol was added to the content of the vial, and it was kept at -20°C for 15 minutes, and then centrifuged in 12000 g resulting in some DNA samples sediment. Then, DNA samples were dried in lab temperature. Finally, dried DNA samples were dissolved in 50 µL of double distilled water (10). Nanodrop and electrophoresis on 1% agarose gel were used to determine the quantity and quality of extracted DNA samples.
3.3. PCR Test to Detect Thermonuclease Gene (nuc)
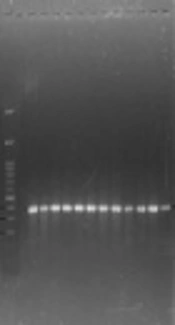
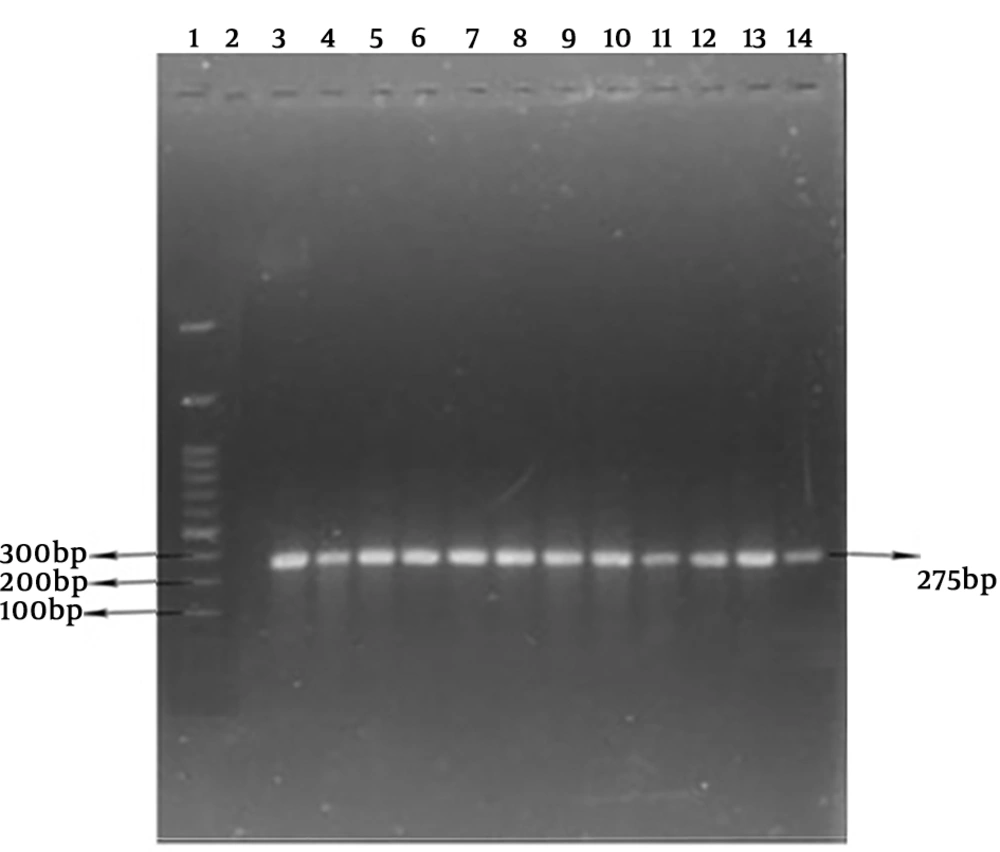
The polymerase chain reaction (PCR) method was done in 25 μL, including Master mix 12.5 mL PCR, 0.4 µM specific primers (Table 1), and DNA extracted containing 1 µL (50 ng). The polymerase chain reaction was performed with primary denaturation cycles at 94°C for four minutes, 32 cycles with denaturation step at 94°C for one minute, the primer annealing step at a temperature of 55°C for one min, extension at 72°C for one minute, and finally, an extension cycle at 72°C for 10 minutes. The PCR production was electrophoresed on 1.5% agarose gel. The extracted DNA of S. aureus PTCC1112 used as positive control and double distilled water was considered as negative control. The isolates were considered as S. aureus after definite diagnosis of nuc existence with a length of 275 bp.
3.4. PCR Test to Identify TSST-1 and mecA Genes
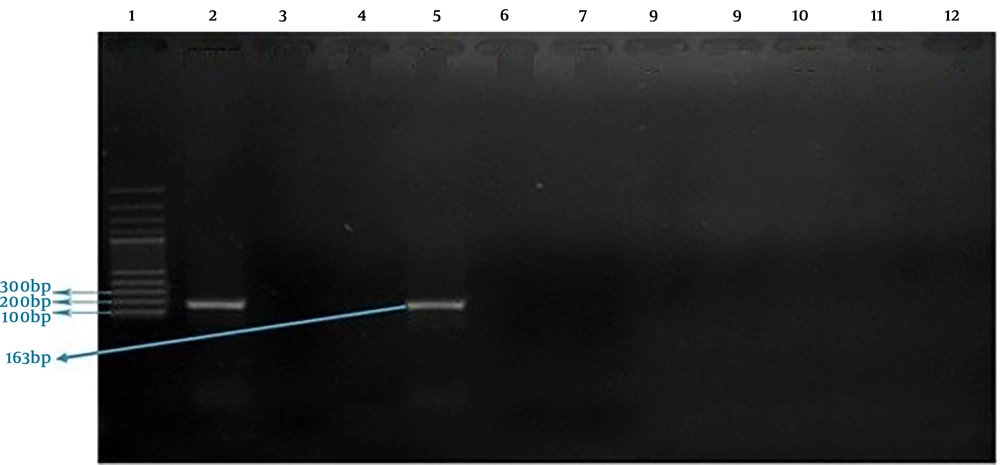
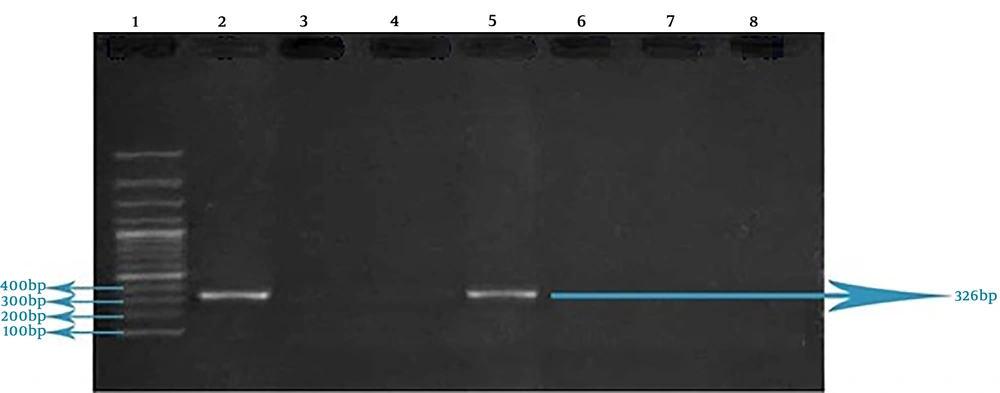
This reaction was performed in a 20 µL volume, which includes 8.5 µL double distilled water, 2 µL buffer 10 X, 2.5 µL dNTPs (10 mM), 1 µL of each one of primers associated with TSST-1 and mecA genes (0.5 µL of forward primer and 0.5 µL of reverse primer, 10 Mm) (Table 1), 2 µL MgCl2 (50 mM), 2 µL Taq DNA Polymerase, and 1 µL extracted DNA. Proliferation of TSST-1 and mecA genes in thermocycler was performed as following: primary denaturation at 95°C for four min, 32 cycles with denaturation stage at 95°C for one min, annealing stage at 52°C for one min, extension stage at 72°C for one min, and finally, a terminal extension stage at 72°C for one min. PCR products related to the proliferation of each one of TSST-1 and mecA genes were evaluated by electrophoresis on 1% agarose gel and 100bp DNA ladder was used to determine the size of fragments. The extracted DNA of S. aureus PTCC1112 used as positive control and double distilled water was considered as negative control.
4. Results
The obtained results showed that all 22 isolated S. aureus strains were positive for the nuc gene (Figure 1).
Of 22 S. aureus isolates, one case (4.54%) contained mecA gene and two cases (9.09%) possessed TSST-1 gene. None of the tested isolates harbored intended genes simultaneously. S. aureus PTCC 1112 (as standard bacterium) didn’t possess the TSST-1 gene (Figures 2 and 3).
5. Discussion
This research reported the frequencies of mecA (4.54%) and TSST-1 (9.09%) genes. Eshraghi et al. (13), reported that 12% of S. aureus samples isolated from foodstuffs in central zones of Iran harbored the mecA gene. Morad Nia et al. (14), reported that 1.6% of S. aureus samples isolated from dairy products in northeast of Iran harbored the mecA gene, which agrees with the finding of the current study. Alizadeh and Amini (10) reported that 60% of S. aureus samples isolated from dairy products in Kerman province in Iran harbored the mecA gene. Pexara et al. (15), showed that the highest prevalence of methicillin-resistant S. aureus (MRSA) in milk and dairy products is found in Ethiopia (60.3%); whereas the MRSA prevalence in Asian countries is variable (for instance, while it is 28.3% in Iran, it is very low in Korea and Japan). The MRSA prevalence in dairy products has been reported to be low in European countries. In addition, the strains prevalence in USA and Canada have been reported as zero to low (15). The MRSA prevalence varies in different countries in the world. This frequency is very low in some countries such as Norway and Netherlands (about 1%) and in some countries, such as India, it is reported between 0% - 10% (16). Study of prevalence of MRSA is one of the ways of assessing hygienic conditions in dairy cattle herds and public health hazards when there are antibiotic resistant strains (15). Arcuri et al. (17), reported that 11.4% of S. aureus samples isolated from Minas frescal cheese in Brazil possessed TSST-1 gene. It has been also reported that the prevalence of TSST-1 producing gene in S. aureus is between 0% - 37% in other parts of the world (18). The researches have shown that TSST-1 is one of the agents involved in causing damage to breast tissue in clinical and subclinical mastitis in cows, sheep, and goats (19). The presence of TSST-1 in domestic animals can be considered as one of the transmission routes to milk and foodstuffs. The researches have shown that 15.5% of S. aureus strains comes from the bovine mastitis possessed TSST-1 gene in the northwest of Iran (20). Studies performed on S. aureus strains originated from dairy products, particularly in endemic strains of Iran, are limited in that in similar studies 66.25% (21) and 15.5% (20) of S. aureus strains originated from local dairy products harbored TSST-1 gene, which is a relatively higher rate when compared to the current research.
Significant differences in the frequency of TSST-1 and mecA genes in different studies may be due to different sources of isolation in different regions, their methods of investigation and sensitivity, and number and types of samples. The results of the current study indicate the difference in dispersion of TSST-1 and mecA genes in S. aureus; this difference probably originates from geographical diversities and also differences in the ecological origin of the isolated strains (milk, human and different animals).