1. Introduction
miRNAs constitute a novel class of small non-coding RNAs consisting of 21 - 25 nucleotides, which play significant roles in the gene expression regulation in more than 60% of all human genes (1-3). miRNAs have also significant roles in manipulating messages transmitted by protein-encoding genes through the inhibition of dynamic translation (3). Recently, many short RNAs have been found to involve in the gene expression regulation and transposon silencing (4). Previous research has declared that lin-4 mRNA is able to repress lin-14 mRNA through matched base pairing in C. elegans (5-7). The principal of the gene expression regulation by miRNA is pairing with their seed sequence to the 3’-UTR sequences matched of the target molecules (8). An alteration in the miRNAs gene expression may be played as an oncomiR or antioncomiR in progression or pathogenesis of hematopoietic disorders (9). Acute lymphoblastic leukemia (ALL) is a heterogeneous disease, consisting of several sub-types with different molecular characteristics. To date, the biological information regarding ALL has widely increased. Determining the gene expression profiles has provided to describe special signatures for various ALL sub-types and allowed for the description of pathways deregulated by damage. Previous microarray analysis revealed that miRNAs play a critical role in several functional categories related to cancer and cell cycle regulation (10). Upregulation of miRNAs can lead to decreased TSG, which is involved in cell progression and differentiation, thereby contributing to ALL formation. In addition, miRNAs could decrease different proteins levels with oncogenic function (11). Earlier investigations have explained that the roles of miRNAs as a TSG in erythroblast cells. In addition, several miRNAs play roles as an oncogene in solid malignancies (12).
2. Micro-RNA Biogenesis and Functions
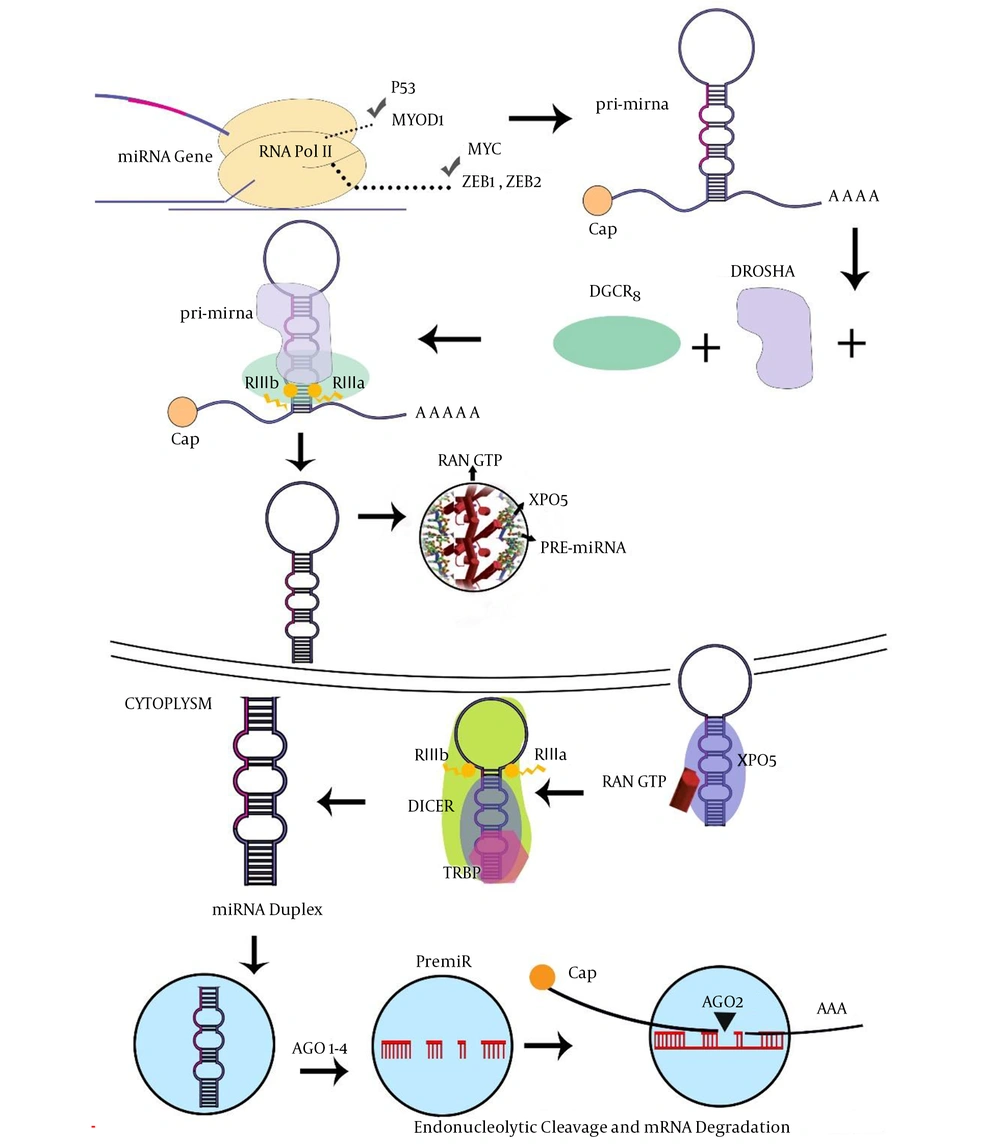
The wide ranges of miRNAs are transcribed within the intron. miRNA is capable of binding to the 3’-UTR sequence of the coding parts of target mRNA. Indeed, the miRNAs molecule, which is generated by RNA Pol-III, has also been suggested for miRNAs encoded within Alu repeat sequences, while the miRNAs are transcribed by RNA pol II, consisting of a 5’-capped in pri-miRNA (13). Several percentages of miRNAs were co-transcribed as gene clusters, encoding various miRNA sequences in a single pri-microRNAs transcript (14, 15). In the next step, pri-miRNAs molecules are processed by the microprocessor proteins, such as Drosha and DiGeorge critical region-8 (DGCR8) cofactor to form pre-microRNAs (16). Then, exportin-5 (XPO5) transfers pre-miRNAs to the cytoplasm with the contribution of RAN-GTPase protein (17). Dicer and TAR-RNA binding protein (TRBP) cofactors create an asymmetric miRNA, miRNA duplex, which involves the miRNA sequence and the antisense miRNA passenger strand (18). Furthermore, this duplex is arranged into the miRNA-induced silencing complex (miRISC) where the protein Argonaute-2 (AGO2) acts as a key effector molecule. The mature miRNA strand is kept by the miRISC complex, whereas the passenger strand is commonly degraded (19). However, the numerous miRNA passenger strands are similarly able to silence target mRNA and possibly have an additional biological role in procedures (Figure 1). Mammalian cells have many miRNAs that correlate with several molecular pathways such as cell cycle regulation, proliferation, apoptosis, differentiation, and cell cycle regulation in hematopoiesis (20). Currently, surveys have highlighted the significant conception of the mechanism of mRNA regulation by investigating variations in miRNA expression in a range of human pathological conditions, consisting of malignancies (21). Given their importance in improvement, miRNAs would also have a powerful aspect in tumorigenesis. Since their identity refers to more than 3000 reports consisting of over 1000 investigations, definite associations between miRNAs and malignancy (22, 23).
Micro-RNA biogenesis and functions (7). Abbreviations: AGO2, protein argonaute-2; DGCR8, DiGeorge critical region 8; TRBP, TAR-RNA binding protein; XPO5, exportin-5.
3. Deregulation of miRNAs in ALL
During the hematopoietic cells differentiation and development, lymphoid and myeloid progenitor cells can lead to leukemia. ALL is a possible derivative of B or T-cell progenitor cells, in which B-ALL is considered as one of the most common malignancies in children. Many studies have reported recurrent cytogenetic abnormalities in B-ALL including the fusion of ETS variant 6 (ETV6) and RUNX1 genes (ETV6-RUNX1), BCR-ABL, mixed lineage leukemia (MLL) rearrangements, and the fusion protein of E2A and PBX1 genes (E2APBX1), which account for about 30 percent of cases (24). Data gathered in various investigations have shown that several miRNA genes are related to ALL. In addition, the long non-coding RNA (lncRNA) is another type of ncRNAs, which plays a critical role in the pathogenesis of cancers (25). Two very important profiling investigations have recently provided a new conception of transcripts, which are complicated in hematopoietic disorders (26). Fang et al. investigated the lncRNA gene expression changes in MLL-rearranged (MLL-r) ALL patient specimens (25), which showed 111 new classes of lncRNAs expressed in MLL-r sub-types of ALL patients. The lncRNAs are related to large groups of genes such as MLL-fusion target proteins, homeobox protein hox-A9 (HOXA9), and MEIS homeobox 1 (MEIS1). The results demonstrated that particular lncRNAs can affect the cellular apoptosis and proliferation through small interfering RNAs (27-31). Collectively, the results indicated a positive association between the abnormal gene expression of B-cell maturation genes and carcinogenesis of B-ALL. In addition, several miRNAs have been identified to be able to create an ALL condition in vivo (31). ALL is a heterogeneous disease defined by numerous fundamental genetic deformities. The rigorous combination of chemotherapy programs has led to an approximately 80% increase in the five-year survival rate of children while in ALL adult patients, it is much lower to obtain successful therapy resulting in nearly a 40% increase in five-year event-free survival (32).
3.1. miR-17-92 Cluster
The miR-17-92 cluster is an example of polycistronic miRNA, which has been most commonly investigated; it is mapped at chromosome 13 and codes several different miRNAs including miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a. The constructions of the cluster gene family are significantly conserved in vertebrates. Initial evolutionary duplication and deletion of genes result in the miR-106b-25 and miR-106a-363 clusters (33). The miR-106b-25 cluster exists on chromosome 7 of the human within intron number 13 on the minichromosome maintenance protein complex (MCM) gene, whereas miR-106a-363 is found on the X chromosome. mir-17- 92 and miR-106b-25 are over-expressed in a wide range of mouse tissues and are predominantly plentiful in embryonic stem cells and during embryogenesis, whereas the miR-106a-363 commonly is downregulated (34-37). Currently, the findings revealed that the miRNA processing pathway could modulate different cellular processes because multiple steps are needed to create mature miRNAs (38-40).
According to the previous investigation, miR-17b92 is controlled through BCR-ABL gene fusion in chronic myelogenous leukemia (CML), as well as ALL malignancies. The copy number of miRNAs encoded by miR-17b92 notably decreased in BCR-ABL positive compared to negative ALL-cells that leads to the promotion of apoptosis in a BCR-ABL dependent mode (41). Previous findings revealed that miR-17b92 directly regulates Bim (Bcl-2-interacting mediator of cell death) and phosphatase and tensin homolog (PTEN) pro-apoptotic molecules in normal lymphopoiesis (42-44). Moreover, miRNA controls Bcl-2 (B-cell lymphoma 2) gene expression and other apoptosis elements, which forms a crucial equilibrium between growth signals and apoptosis in ALL patients with BCR-ABL-positivity (45-47). Although the genetic status of ALL subtype can influence the outcome of children with ALL, the cellular drug resistance plays a critical role in the success of treatment (48-50). Knowledge of the different expression levels of miRNA in different genetic subgroups of ALL can help a better understanding of the biology of the genetic abnormalities and drug resistance in these patients.
3.2. miRNA-150
miR-150 has been studied in many analytic investigations and it was discovered to have possible prognostic importance. miR-150, located on chromosome 19q13, has demonstrated as a hematopoietic-specific miRNA in lymphoma. In CML, it has been declared to relatively decrease the levels of gene expression in patients with negative clinico-biological indicators and poorer prognosis (51). During B and T cells maturation, miRNA-150 was expressed to play crucial roles in the transition from progenitor B cell to the precursor B cell stage. Premature expression of miRNA-150 can lead to inhibition of the progenitor B transfer to precursor B cell (52-54). In the thymus, the expression of miRNA-150 may promote T cell development (55). One of the targets of miRNA-150 is c-myb, which is involved in early lymphoid development as an essential transcription factor.
3.3. miR-155
miR-155 is produced from ncRNA transcribed from the B-cell integration cluster, a gene situated on chromosome 21. miR-155 is a multiple function miRNA, which has been identified to be associated with different biological pathways. miR-155 is known to be a promoter of many different types of cancers such as lymphomas. miR-155 plays important roles in many different cellular pathways including an integral role in numerous pathways signaling for ATM, ERK/ MAPK, SAPK/JNK, Toll-like receptors, and the B-cell receptors (56). MiR-155 also targets anti-apoptotic factors and promotes apoptosis (57).
3.4. miR-181a
A negative correlation between miR-181a and early growth response protein 1 (EGR1) has been observed, indicating EGR1 is a direct target for miR-181a so that the decrease of EGR1 expression was shown when miR-181a was over-expressed. miR-181a overexpression in Jurkat T-ALL cells increases cell proliferation and progression from G1 to S phase during the cell cycle. EGR1 has the role of TS in ALL and miR-181a acts as an oncomiR in ALL patients through decreasing EGR1 (58). Other investigations are needed to more specifically evaluate the role of the miR-181a/EGR1 interaction. However, similar investigations will open new windows for future diagnosis, prognosis, and therapeutic target in patients with ALL (58).
3.5. miR-708-5p
miR-708-5p was early reported in cancerous and non-cancerous cervical cases and has a considerable sequence resemblance to miR-28 (59, 60). miR-708-5p is deposited on chromosome 11 (11q14.1) as a mirtron, a microRNA encoded within an intron of a protein-coding gene. miR-708-5p is located within intron 1 of the ODZ4 gene, which encodes the transmembrane protein Teneurin Transmembrane Protein 4 (61). Studies using quantitative PCR have shown that miR-708 is differentially expressed in ALL subtypes. The finding revealed that miR-708 was upregulated in the B-cells group as compared to normal CD34 B cells. It is suggested that miR-708 remarkably correlates with the leukemic subtype compared to the maturation status of cells (62).
3.6. miR-29a
Previous findings described that miR-29a is one of the early miRNAs with the strength to both inhibit and increase tumor advancement depending on the oncogenic context. Earlier investigations have reported that the down-regulation or deficiency of miR-29 family members and up-regulation of their oncogenic targets such as Tcl1, Mcl1, and DNMT3 have been affected in several malignancies (63-65). According to the findings revealed by Oliveira et al., it was proposed that decreased miR-29a levels may move to the altered epigenetic condition of T-ALL, asserting its usefulness in the physiopathology of this complication (66, 67).
3.7. miRNA-192
First, Lim et al. explained miR-192 as a novel diagnosed miRNA, which was co-transcribed with miR-194 (68). Several investigations described the overexpression of miR-192 in various cancer types, consisting of gastric malignancy, hepatocellular disease, and RB (69). Conversely, miR-192 was downregulated in colorectal cancer (CRC) and hematological disorders, as well as in ALL where it was correlated with weak prognosis.
4. Conclusions
The miRNA expression alterations can result in insufficient onco-miR or TS-miR expression and lead to the development of leukemia. Up-regulation or down-regulation of several miRNAs such as miR-17-92 cluster, miRNA-150, miR-155, miR-181a, miR-708-5p, miR-29a, and miRNA-192 may be considered of clinical importance. Furthermore, it is conceivable to predict new functions for ncRNAs and they might be implicated as diagnostic or prognostic biomarkers in different classes of leukemia such as ALL.