1. Background
Salmonellae are non-sporulating, flagellated, Gram-negative, facultative anaerobic bacilli belonging to the Enterobacteriaceae family containing more than 2,300 serotypes. Antibiotics such as ampicillin, chloramphenicol, and trimethoprim/sulfamethoxazole have been used for the treatment of enteric fever for many years. Later in the 1980s, the resistance of Salmonellae towards multiple drugs was reported. This caused the increasing use of fluoroquinolone alternatives such as ciprofloxacin and extended-spectrum cephalosporins (1). Recently, azithromycin has also been reported to be effective in the treatment of enteric fever (2, 3).
The literature highlights the production of extended-spectrum β-lactamase (ESBL) in Salmonella typhi, which is directly related to their antibiotic resistance. This resulted in the failure of treatment in many countries where cefotaxime, ceftriaxone, and ciprofloxacin were used as antibiotics (4).
The ESBLs reported in Salmonella spp. include TEM (Temoneira), SHV (containing sulfhydryl variable active sites), and CTX-M (cefotaximase hydrolyzing activity) (5). The most commonly found ESBLs in Enterobacteriaceae, including Salmonellae, are CTX-M. In 1989, the earliest CTX-M1 type of cefotaximase was isolated in Germany (6). Around 300 different types of ESBLs have been reported (7), with TEM and SHV being the most commonly occurring variants (8). Nevertheless, over the past decade, several countries have witnessed a rise in non-TEM, non-SHV variants of ESBLs such as CTX-M (8). ESBL generation is mainly linked to mutations in β-lactamase enzymes encoded by blaSHV, blaTEM, and blaCTX-M genes.
2. Objectives
The present study aimed to document the existing dominance of ESBL-producing Salmonella spp. found in poultry meat from the commercial broiler (CB) and spent hens (SH) in Ardabil, located in the northwest of Iran.
3. Methods
This study was conducted in the Department of Microbiology of Ardabil University. The following materials and methods were used:
3.1. Bacterial Isolation
Overall, 100 retail chicken carcasses (comprising 50 from the CB and 50 from the SH) were used to collect test meat samples. The samples were taken from the CB and SH of local supermarkets and wet markets of Ardabil city. In total, 20 Salmonella strains were isolated from these collected meat samples. To this end, 25 g of each sample was transferred to 225 mL of the pre-enrichment medium consisting of peptone water and placed in an incubator at 37°C for 24 hours. The broth was then added to the selenite-F environment and incubated for 24 hours. The selenite-F medium was then inoculated into Salmonella-Shigella agar and brilliant green agar media and incubated for 24 hours at 37°C. Colorless colonies with black centers appeared in Salmonella-Shigella agar and yellow-colored colonies with black centers were visible in the brilliant green agar. These were suspected to be the colonies of Salmonella and were identified by using several biochemical tests such as urea, IMVIC, and TSI. Isolates enriched in selenite-F broth were carefully streaked on Salmonella-Shigella agar plates and incubated. Pure colonies were selected from the agar plates and then identified with standard bacteriological techniques.
3.2. Serotype Identification
The Kauffmann-White scheme was used for the identification of Salmonella serotype based on surface antigens. Serotyping was performed after the identification of species on a fresh pure culture of Salmonella. The antisera of Salmonella were purchased from Padtan-Teb Company. With the help of polyvalent antisera, the agglutination test was performed. The determination of serogroup was done by the agglutination on a slide with O-somatic antiserum. Then, specific monovalent antisera were used and allowed to stand on it for 10 seconds so that the antigen could be identified. The primary culture was treated with saline and then with O-antigen. The type of O-antigen was determined by oligosaccharides associated with outer membrane lipopolysaccharides. If agglutination occurred, it indicated that the serotype had been treated with a specific antiserum. After this, the serotype was treated with H-antigen (phase 1 and phase 2). The type of H-antigen was determined by the flagellar protein of bacteria. Based on the agglutination pattern obtained after this procedure, the serotype was identified by the Kauffmann-White reference catalog as B or D. In the Kauffmann-White (KW) scheme, the antigenic properties and variations of the O (surface polysaccharide) and H (flagellar) antigens from each serovar are described, which are known as the antigenic formulae.
3.3. Antimicrobial Susceptibility
Using the disk diffusion technique, the susceptibility of Salmonella towards antibiotics was determined on Mueller-Hinton agar plates. Interpretations were made in accordance with the guidelines of CLSI 2015 (9). The antibiotics tested were amoxicillin (30 µg), ampicillin (10 µg), cefotaxime (30 µg), ciprofloxacin (5 µg), nalidixic acid (10 µg), spectinomycin (10 µg), tetracycline (30 µg), and sulfamethoxazole (1.25/23.75 µg). The minimum inhibitory concentration (MIC) of cefotaxime and ceftriaxone was determined by the agar dilution method with the tested range of 0.008 - 128 μg/mL and then interpreted in accordance with the CLSI 2015 guidelines (9). In order to determine the MIC, different intended concentrations of each antibiotic were inoculated by using a fixed volume of nutrient broth containing a standard concentration of bacteria (0.5 McFarland) and then, the suspensions were incubated and examined for turbidity. A turbid sample indicated bacterial growth whereas a clear sample indicated the inhibition of bacterial growth and the corresponding concentration of the antibiotic used was considered as the MIC.
3.4. Screening for ESBL Production
The screening of ESBL production in Salmonella isolates was performed by using the inhibition zone test in accordance with the NCCLS recommendations. Discs of ceftazidime (30 µg) and cefotaxime (30 µg) were used for this study.
3.5. Phenotypic Confirmatory Test for ESBL Production
Mueller-Hinton agar culture medium was used to perform this study. Four different antibiotic discs were used, which included cefotaxime (30 µg), cefotaxime/clavulanic acid (30 µg/10 µg), ceftazidime (30 µg), and ceftazidime/clavulanic acid (30 µg/10 µg). The study was performed in accordance with the NCCLS recommendations.
3.6. Polymerase Chain Reaction (PCR) Amplification
The bacterial DNA was extracted by using the boiling method (10, 11). From tryptic soy broth (TSB), Salmonella strains were reactivated on tryptic soy agar (TSA) upon incubation for 18 - 24 hours. Later, they were inoculated in Luria Bertani broth (LB broth, 2 mL). After incubation for 18 - 24 hours, the LB broth was centrifuged (10,000 rpm for 10 minutes). The bacterial cell pellet collected was re-suspended in 500 μL of phosphate buffer (100 mM, pH 7) to enfeeble the cell membranes. Upon immersion of re-suspended bacterial cells in a boiling water bath (100°C) for 15 minutes, the membranes ruptured to release the genetic material. The solution containing the genetic material was collected and the DNA content was precipitated with 250 μL of absolute alcohol; then, it was washed twice with 1000 μL of 70% alcohol (stored at -20°C), dried, and re-suspended in 100 μL of sterile water.
This method was used for preparing lysates for all the isolates. All the isolates were tested positive for ESBLs production phenotypically. They were later screened for the presence of blaCTX, blaSHV, and blaTEM genes by PCR assay using specific primers listed in Table 1. The PCR assay was carried out in 0.2-mL thin wall tubes using each of the bacterial lysates as template DNA. Each tube consisted of 1.5 mM MgCl2, 0.2 µM of each primer, 200 µM of each dNTPs, 1.5 U of Taq polymerase (CinnaGen, Tehran, Iran), and 2.0 µL DNA template, making up overall 25 µL as the final volume. The PCR assay was performed in a thermal cycler and the cycling condition for blaCTX-M-1 and blaSHV was maintained as follows: (I) Initial denaturation at 94°C for seven minutes; (II) 30 cycles of amplification with denaturation at 94°C for 30 seconds; (III) annealing at 57°C for 30 seconds and an extension at 72°C for 30 seconds; and (IV) a final extension at 72°C for five minutes. For the blaTEM gene, the annealing temperature was maintained at 53°C. The multiplex PCR assay was performed using the same composition of the PCR mixture described earlier, with an annealing temperature of 54°C.
| Gene | Primer Sequence | Product Size | Reference |
|---|---|---|---|
| blaCTX | 950 | Schmitt et al. (12) | |
| Forward | 5-CCCATGGTTAAAAAACACTGC-3 | ||
| Reverse | 5-CAGCGCTTTTGCCGTCTAAG-3 | ||
| blaTEM | 1080 | Weill et al. (13) | |
| Forward | 5-ATAAAATTCTTGAAGACGAAA-3 | ||
| Reverse | 5-GACAGTTACCAATGCTTAATC-3 | ||
| blaSHV | 747 | Sun et al. (14) | |
| Forward | 5-ATGCGTTATATTCHCCTGTG-3 | ||
| Reverse | 5-TGCTTTGTTCCGGGCCAAAC-3 |
The Pattern of blaTEM, blaSHV, and blaCTX Genes
4. Results
20 Salmonella strains were isolated from 100 poultry meat samples collected. 55% of the isolates (11/20) were obtained from CB meat and the remaining 45% (9/20) from SH meat. The isolated Salmonella serogroups were identified as B (five isolates) or D (15 isolates). The results are provided in Table 2. The antibiotic susceptibility test showed a significant amount of antibiotic resistance. The phenotypic confirmatory test was used for the detection of β-lactamases enzymes. An increase of > 5 mm in the inhibition zone diameter was observed for all the antimicrobials when used in combination with clavulanic acid compared to when used alone. This is possibly due to the pronounced effect of ESBL on the individual antimicrobials compared to their combination with clavulanic acid. The size of the inhibition zone was considered as a marker indicating the presence or absence of ESBL in pre-screening. The positive indication for the presence of ESBL was given by the inhibition zone diameters obtained from the antibiotics, i.e. the inhibition zone diameter of ≤ 22 mm for ceftazidime and ≤ 27 mm for cefotaxime, further analyzed for the ESBL presence by the ESBL confirmatory test.
The results of the initial screening test were in accordance with the results obtained from the complementary confirmatory test of ESBL, with more than 90% of the isolates being positive in both analytical tests. The negative results of the screening test were also in agreement with the confirmatory test results, with some exceptions. For a few Salmonella isolates, although a positive outcome was obtained in the screening test, the confirmatory test result for ESBL production was found to be negative. Thus, the overall results indicated that the routine susceptibility test is a relatively less sensitive technique, failing to identify ESBL production in 25% of B (Salmonella typhimorium) and 75% of D (Salmonella enteritidis) isolates. The results are summarized in Table 2.
| Serotypes | B (Salmonella typhimurium) | D (Salmonella enteritidis) |
|---|---|---|
| No. of isolates | 5 | 15 |
| ESBL producers, % | 25 | 75 |
The Results of Serotype Identification and ESBL Production Obtained from the Phenotypic Confirmatory Test (Total No. of Isolates, N = 20).
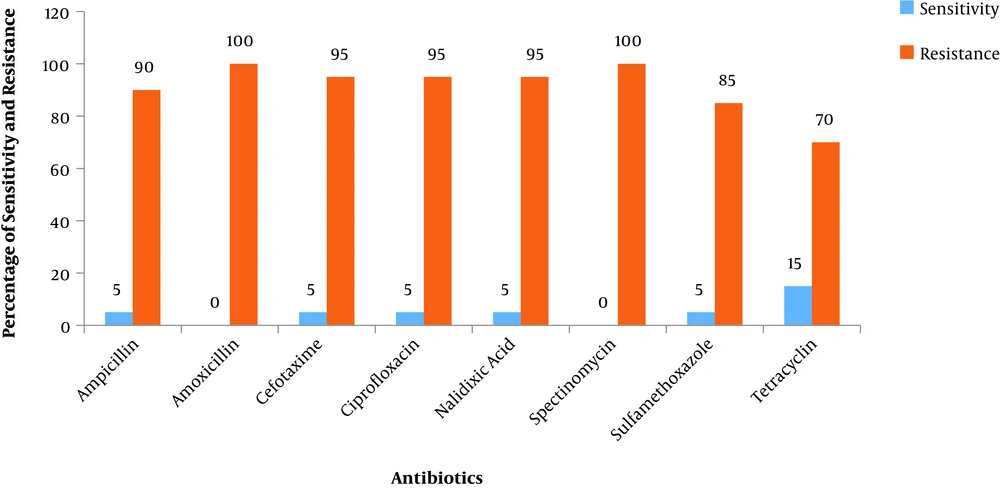
In fact, the results obtained from the routine susceptibility test showed that Salmonella strains belonging to these two serotypes exhibited antibiotic susceptibility towards cefotaxime and ceftazidime to some extent. About 78.7% - 94.0% of the total isolates that were cephalosporin-resistant were found to produce ESBL. The results are graphically represented in Figure 1 where the percentage of Salmonella isolates (out of 20 isolates) exhibiting susceptibility or resistance towards each antibiotic has been plotted. The analysis of antimicrobial-resistance showed that resistance to β-lactam agents was more frequent in ESBL-producing isolates (Figure 1).
Percentage of Salmonella isolates (total of 20 isolates) exhibiting susceptibility or resistance towards each of the following antibiotics: amoxicillin (30 µg), ampicillin (10 µg), cefotaxime (30 µg), ciprofloxacin (5 µg), nalidixic acid (10 µg), spectinomycin (10 µg), tetracycline (30 µg), and sulfamethoxazole (1.25/23.75 µg).
Figure 1 clearly shows that, in general, Salmonella spp. isolates are resistant towards most of the antibiotics. The isolated samples of Salmonella from both CB and SH showed the significant presence of both β-lactamases enzymes. Moreover, all the Salmonella isolates showed a high antibiotic resistance to amoxicillin (100%), spectinomycin (100%), cefotaxime (95%), nalidixic acid (95%), and ciprofloxacin (95%). The lowest resistance was shown to tetracycline (70%) as observed in Table 3.
| S. No. | Antibiotics | Susceptibility, % | Resistance, % |
|---|---|---|---|
| 1 | Ampicillin | 5 | 90 |
| 2 | Amoxicillin | 0 | 100 |
| 3 | Cefotaxime | 5 | 95 |
| 4 | Ciprofloxacin | 5 | 95 |
| 5 | Nalidixic acid | 5 | 95 |
| 6 | Spectinomycin | 0 | 100 |
| 7 | Tetracycline | 5 | 85 |
| 8 | Sulfamethoxazole | 15 | 70 |
Susceptibility and Resistance of Different Antibiotics (in Percentages) Against Salmonella spp.
The results of PCR amplification of blaTEM, blaCTX-M, and blaSHV genes in Salmonella isolates from the CB and SH are given in Figure 2.
Agarose gel electrophoresis of the PCR amplified products. M: Molecular ladder; 1: blaTEM positive control at 1080 bp; 2: Salmonella isolates showing bands at 1080 bp and 950 bp; 3: Negative control; 4: blaCTX positive control at 950 bp; 5: Salmonella isolates showing bands at 1080 bp and 747 bp; 6: blaSHV positive control at 747 bp; and 7: Salmonella isolates showing a band at 1080 bp only.
blaTEM was the dominant β-lactamase gene (85%), followed by blaCTX (60%) and blaSHV (35%) (Table 4). The accession numbers JQ735915 for the β-lactamase blaTEM gene, JN003854 for the blaSHV gene, and EF592571 for the blaCTX-M gene from GenBank were used. In addition to the study for the detection of β-lactamases, it was also revealed that a significant number of antibiotic-resistant Salmonella were presently isolated from the retail poultry meat samples of CB and SH.
| Genes | No. of Isolates (Frequency%) |
|---|---|
| TEM | 17 (85) |
| CTX-M | 12 (60) |
| SHV | 7 (35) |
The Total Number of Genes Amplified During PCR (Total Number of Isolates, N = 20)
5. Discussion
Enteric fever is endemic due to Salmonella enterica. The pathogens are transmitted through the fecal-oral route due to the lack of hygiene. Antibiotic therapy constitutes the mainstay of the management of enteric fever. The purpose of the current study was to detect β-lactamase enzymes produced by Salmonella isolated from poultry meat through two advanced detection procedures comprising phenotypic and molecular detection. 20 Salmonella isolates were obtained from 100 meat samples, with 11 isolates from the CB and nine isolates from the SH. The serotypes identified were B (Salmonella typhimurium, five isolates) and D (Salmonella enteritidis, 15 isolates). The phenotypic detection of metallo-β-lactamase was done in 11 (100%) CB and nine (100%) SH samples.
In the present study, the susceptibility of Salmonella was studied against antibiotics such as amoxicillin (30 µg), ampicillin (10 µg), cefotaxime (30 µg), ciprofloxacin (5 µg), nalidixic acid (10 µg), spectinomycin (10 µg), tetracycline (30 µg), and sulfamethoxazole (1.25/23.75 µg). Salmonella spp. isolates were found to be resistant towards most of the antibiotics, with very less susceptibility to tetracycline and ampicillin. In a similar study, Bradford (15) reported the least antibiotic resistance of Salmonella isolates towards aztreonam (10%), followed by ceftazidime (14%), ceftriaxone (21%), cefoxitin (23%), and cefotaxime (32%). In addition, ESBL producers offered higher antibiotic resistance compared to non-ESBL producers. A relationship was observed in Salmonella spp. between ESBL production and antibiotic resistance towards ceftazidime and ceftriaxone. This reinforces the recent finding that most ESBLs display a special affinity to degrade ceftazidime (16). In this study, the 20 isolates obtained from the CB and SH were analyzed using PCR assay for the simultaneous detection of blaSHV 747 bp, blaTEM 950 bp, and blaCTX 1080 bp. Ehlers et al. (16) found that M-PCR simultaneously amplified and detected the presence of blaSHV (747 bp), blaCTX-M (593 bp), and blaTEM (445 bp). In a study carried out by Qiao et al. (17), it was found that 57.3% of the isolates harbored blaTEM whereas 30.2%, 24.0%, 18.8%, and 7.3% of the isolates carried blaOXA-1, blaCTX-M-15, blaCTX-M-3, and blaPSE-1 genes, respectively. In our study, the comparatively higher values of 85%, 60%, and 35% were found for TEM, SHV, and CTX-M genes, respectively. These results are consistent with the findings reported from Tunisia where 86.44% of ESBL producing Klebsiella spp. were isolated (18). These findings show the importance of detection of resistant genes to ensure the appropriate treatment and use of proper control measures against such infections and their causative agents.
The underlying mechanism of β-lactam antibiotic resistance is through the production of β-lactamases. These enzymes function by hydrolyzing the β-lactam ring by breaking the amide linkage, thereby disabling their capability of inhibiting bacterial cell wall synthesis (19-21). Previously, it was known that ESBL-producing bacteria thrived commonly in hospital and clinical settings where extensive use of antimicrobial drugs and agents assisted in the development of their antimicrobial resistance (22). However, in the present study, we report the isolation and identification of ESBL isolates from poultry meat from broiler production chain. This study has a serious implication and intends to draw attention to the fact such pathogenic microorganisms can easily find a way into the human food chain (23). Moreover, such pathogens not only can spread from infected chicken to all other meat products, but also can contaminate an entire slaughter line triggering an outbreak of food poisoning (24-27).
The emergence of cheap generic alternatives has allowed the continued usage of prohibited drugs and additives in poultry farming as common measures to prevent infection. In addition, the extensive use of prescribed β-lactam antibiotics to treat Salmonella infections has contributed to a rise in antibiotic-resistant microorganisms (28, 29).
5.1. Conclusions
In this study, the isolated Salmonella were identified as B (five isolates) or D (15 isolates) serotypes. The ESBL screening study indicated the presence of ESBL in isolates from both CB and SH. Some of the Salmonella serotypes examined in this study showed resistance to multiple β-lactam antibiotics. An increase in the inhibition zone diameter was observed for all the antimicrobials when used in combination with clavulanic acid compared to when used alone. This is possibly due to the pronounced effect of ESBL on the individual antimicrobials compared to their combination with clavulanic acid. This indicates the scope of combinatorial antibiotic therapy in treating infections caused by Salmonella.