1. Background
Colorectal cancer (CRC) is the third adenocarcinoma type, which has a high prevalence and mortality rate worldwide. Fifty percent of patients with CRC are diagnosed in the last stage of disease (1). The molecular pathogenesis of CRC is complicated and comprised of several regulatory processes such as gene rearrangements, chromatin remodeling, epigenetic, genetic alterations, and gene expression alterations (2-6). It is believed that oncogenes and tumor suppressor genes (TSG) are related to the molecular pathogenesis of CRC (7-9). Nevertheless, the exact molecular genetic mechanism of CRC tumorigenesis process remained unclear (10). A novel biomolecular prognostic and diagnostic marker is required to verify the molecular carcinogenic mechanisms and improvement of medicinal protocols in the CRC management (11). A previous investigation performed on the esophageal squamous cell carcinoma (ESCC) tissues showed that a gene copy number was increased in the 5p15.2 region (12). Two novel mRNAs have been investigated in the ESCC tissues and other types of malignancies, which was located in the 5’ upstream of the Delta-catenin gene (2). The JS-2 is one of the novel genes, which was analyzed using qReal-Time PCR method. The findings revealed that the JS-2 gene was over-expressed in 18% of ESCC tissues (11).
The JS-2 encodes a special protein containing 233 amino acids, which does not indicate the amino acid sequence compared with other recognized protein sequences in Swiss-Prot (http://au.expasy.org/sprot/). Although the overexpression of the JS-2 gene was observed in the ESCC and CRC cancerous tissues, future studies are required to reveal the potential clinical roles of JS-2 gene expression level in other human disorders (11, 12).
2. Objectives
Given the JS-2 gene expression pattern has not been effectively explored in different human malignancies, we aimed to investigate the potential role of the JS-2 in patients with CRC. This is the first study to investigate the JS-2 gene expression level in North Western Iranian patients with CRC.
3. Methods
3.1. Specimens Selection
In this case-control study, we evaluated 100 fresh colorectal tissue samples, including 50 cancerous adenocarcinoma and 50 adjacent non-neoplastic tissues, which were obtained from of the patients with CRC undergoing colorectal surgery or colonoscopy, who had been referred to the Imam Reza Hospital, Tabriz University of Medical Sciences, from April 2015 to March 2016. The inclusion criteria consisted of the patients that their malignancy was diagnosed and confirmed by the histopathological tests. The pathological board re-assessed all specimens and determined grades and classification according to the World Health Organization (WHO) criteria (13). The exclusion criteria were the subjects who underwent chemotherapy and radiotherapy regimen before the surgery or colonoscopy. In addition, we excluded the patients with CRC having other malignancies, systemic diseases, and diabetes disorders. Ethics and Human Rights Committee in the pathology center approved the current research, and all subjects signed written informed consents.
3.2. RNA Isolation and Complementary DNA Synthesis
All tissue specimens transferred into the RNA later solution (Qiagen, Germany) and stored at -80°C before RNA isolation. Total RNA was purified from tissues using TRIzol reagent (Molecular Research Center Inc., Cincinnati, OH, USA) and treated with DNase I enzyme (Thermo Scientific™, USA), according to the guideline. To confirm the quality and quantity of separated RNA, we have used 1% agarose gel electrophoresis and NanoDrop instruments (Thermo Scientific™, Wilmington, Denmark). Reverse transcription reaction was performed using 1 µg of total RNA in a 20 µL final reaction volume. RNA was converted to cDNA using cDNA Synthesis Kit (Fermentas, Burlington, Canada) with random hexamer primer, according to the manufacturer’s protocol. All cDNA samples were separately diluted to 50 ng/µL in order to obtain a uniform concentration specimen for qReal-Time PCR experiments. Conventional PCR was done with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers to ensure the proper synthesis of cDNA samples.
3.3. Quantitative Real-Time PCR
Amplification was carried out in the triplicate form using specific primer pairs (11) (Table 1) and SYBR Green-I dye Master Mix (Bioneer, Korea) by the Rotor-GeneTM 6000 systems (Corbett Research, Australia) according to the manufacturer’s guidance. The GAPDH gene was used as the housekeeping gene to normalize gene expression. In all reaction, we included 150 ng cDNA, SYBR Green-I dye Master Mix, and 10 pM of each primer for JS-2 and GAPDH in a 25 µL final volume. Real-time PCR conditions were started with an initial denaturation step (94°C, 5 minutes), followed by 35 cycles of denaturation (94°C, 10 seconds), annealing temperatures (59°C for JS-2 and 62°C for GAPDH) for 10 seconds, followed by extension cycle at 72°C for the 20 seconds. The 2-ΔΔct method was used to measure the relative quantification of gene expression changes.
| Genes/Status | Primers Sequence | Amplicon Size, bp |
|---|---|---|
| JS-2 | 136 | |
| Forward | 5’-ATGCACGAGTTATTGCTTGC-3’ | |
| Reverse | 5’-ATCGATGTACAGGGCCTCTT-3’ | |
| GAPDH | 152 | |
| Forward | 5’-AGGGCTGCTTTTAACTCTGGT-3’ | |
| Reverse | 5’-CCCCACTTGATTTTGGAGGGA-3’ |
Primer Sequences of JS-2 and GAPDH Genes Used in qReal-Time PCR
3.4. Statistical Analysis
The statistical analysis was performed using SPSS version 25.0 (SPSS Inc., Chicago, IL, USA). To compare the JS-2 gene expression change between cancerous and adjacent non-cancerous tissues in subjects with CRC and clinical features, the student t-test and chi-square test were used. The P < 0.05 was considered statistically significant.
4. Results
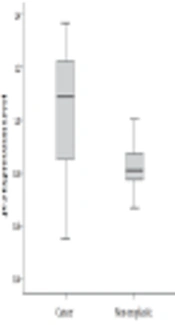
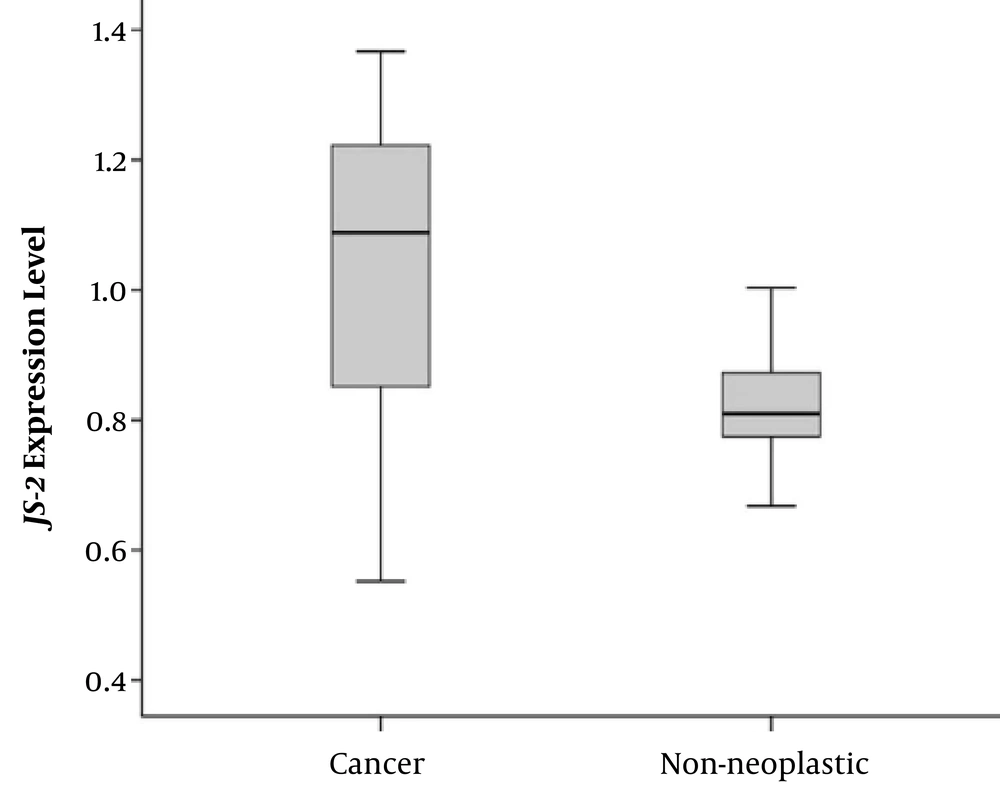
The mean age of the patients with CRC was 50.38 ± 6.12 years (ranging between 45 and 74 years), consisting of 33 (66%) males and 17 (34%) females. We evaluated the JS-2 gene expression change whether significant differences are found between cancerous and non-cancerous tissues of patients with CRC. Our findings showed that the JS-2 gene was over-expressed in cancerous tissues compared with the adjacent non-cancerous ones (P = 0.037), (Figure 1).
In addition, we evaluated the correlation between JS-2 gene expression and clinical parameters in patients with CRC. To assess the correlation between JS-2 gene expression and CRC clinicopathological characteristics, we divided 50 CRC samples into high and low expression according to median Δct (Table 2). The median amount ≥ 0.93 of JS-2 gene expression was considered to be the over-expression group and the median amount < 0.93 considered to be the low-expression group. Overexpression of JS-2 gene was associated with the tumor differentiation (P = 0.047) and tumor stage (P = 0.036), (Table 2). However, the JS-2 expression did not correlate with other characteristics such as age (P = 0.774), sex (P = 0.520), tumor site (P = 0.696), and tumor size (P = 0.851), (Table 2).
| Clinical Parameters | JS-2 Gene Expression Change | P Value | |
|---|---|---|---|
| Low Expression Median Amount < 0.93 | High Expression Median Amount ≥ 0.93 | ||
| Gender | 0.520 | ||
| Male | 7 (14) | 26 (52) | |
| Female | 5 (10) | 12 (24) | |
| Age | 0.774 | ||
| < 50 | 11 (22) | 10 (20) | |
| > 50 | 14 (28) | 15 (30) | |
| Tumor site | 0.696 | ||
| Colon | 15 (30) | 13 (26) | |
| Rectum | 13 (26) | 9 (18) | |
| Tumor size, mm | 0.851 | ||
| < 50 | 15 (30) | 20 (40) | |
| > 50 | 6 (12) | 9 (18) | |
| Histological grade | 0.047 | ||
| Well | 2 (4) | 36 (72) | |
| Moderate + poor | 3 (6) | 9 (18) | |
| T staging | 0.036 | ||
| T1 + T2 | 6 (12) | 25 (50) | |
| T3+ T4 | 9 (18) | 10 (20) | |
The Correlation Between of JS-2 Gene Expression Change and Clinicopathological Features of CRC Patientsa
5. Discussion
This is the first study to evaluate the JS-2 gene expressions change in Iranian patients with CRC. Consistent with other studies, the results of the current study revealed that the JS-2 gene expression alteration may be implicated in the pathogenesis of CRC (11). The findings showed a significant difference in the JS-2 gene expression pattern between cancerous tissues compared with the adjacent non-cancerous tissues (11).
Previous studies reported a new section at the 5p15.33 cytogenetic region, which contains JS-2 gene, involving in the different malignancies such as bladder, thyroid, ESCC, and lung cancer (14-17). Fatima et al. investigated the mRNA expression levels of JS-2 in 20 cases of ESCC and revealed that it was upregulated and downregulated in 14% and 9% of cases, respectively (18). Lam et al. (11) investigated the JS-2 gene expression in 176 colorectal tissues consisting of 116 Adenocarcinomas, 30 adenomas, and 30 non-neoplastic tissues as well as two colorectal cancer cell and benign cell lines, including SW480 and SW48.
The findings revealed that the relative gene expression levels of JS-2 were increased in mucinous adenocarcinoma compared with the non-mucinous adenocarcinoma. In addition, colorectal cancer cell line showed a lower expression of JS-2 compared with the benign cell line (11). The results suggested that the JS-2 gene was upregulated and may have a profound role in the pathogenesis of CRC (11, 18). In addition, Bohm et al. described several TSGs located in 5p13-12 region, which were implicated in carcinogenesis of the solid tumors and were inactivated during tumorigenesis (19). Although the findings of several investigations revealed that the JS-2 gene expression changes involved in the development of different malignancies, the precise roles remained unclear in CRC carcinogenesis (14-17).
These data proposed that the JS-2 gene was over-expressed in the higher advanced tumors without affecting the healthy tissue. However, since JS-2 plays an anti-growth activity, it is conceivable that the JS-2 gene over-expression in higher advanced carcinomas may be a feedback to intensify signaling for TSG arising from gene control processes recognizing the growing malignant cells. In the present investigation, JS-2 gene expression change correlated with the histological grade and T staging of clinical features. However, our findings are compatible with the other results reported and may be implicated in CRC pathogenesis (11).
In conclusion, our findings revealed that the JS-2 gene may be involved in the progression of CRC. In addition, we suggested that the JS-2 gene may be implemented as an important potential prognostic biomarker for CRC patients.