1. Background
Breast cancer is the most frequent cancer and the leading cause of cancer death globally among women (1). Also in Iran, breast cancer is one of the most common cancers in women and 24.4% of all diagnosed cancers in women are breast cancer, which causes more than 1063 deaths annually (2). Because of a lack of definitive treatment to deal with this cancer, early and accurate diagnosis of the disease is critically important because it has a beneficial effect on reducing mortality in patients.
Epigenetic modification refers to both heritable changes in gene activity and expression and chromatin structure that occur without an alteration in the DNA sequence (3). DNA methylation is an epigenetic modification that directly affects DNA, which is the result of transfer of a methyl group from S-adenosyl-methionine (also known as SAM) to the cytosine. CpG islands (The CG island are regions with a high frequency of CpG sites and ‘p’ simply shows that ‘C’ and ‘G’ are connected by a phosphodiester bond) are considered to be a hallmark of the promoter regions of the genes. Extensive genomic analysis has shown that approximately 56% of the human promoters are associated with CpG islands. DNA methylation regulates gene expression with the effect on the upstream region of the gene (4). Changes in the pattern of methylation can lead to tumorigenesis (5).
The RASSF1A (Ras-associated domain family 1) tumor suppressor gene is located on 3 p21.3, the short arm of chromosome 3, which acts as an inhibitor of the cycline D1 protein. Also, it plays a key role as an interactor with the proteins of the retinoblastoma family and negative regulator of cell proliferation through the G1/S inhibitor and mitotic checkpoint (6-10). Loss of expression of this gene occurs more frequently by hypermethylation that is capable of causing the development of human cancers (10).
2. Objectives
The aim of this study was to compare the DNA methylation pattern in the promoter region of the RASSF1A gene between cancerous tissues in the patients affected by breast cancer and normal tissues in the control group in Khuzestan province.
3. Methods
3.1. Study Subjects
In the present study, 20 women were recruited from the Shafa and the Imam Khomeini hospitals in Ahvaz. Written informed consent was obtained from all participants for their tissues to be utilized for this study. Furthermore, twenty ages matched healthy women that had no history of cancer and autoimmune diseases were selected as the controls group. In the control group, the tissue of people who had come for cosmetic surgery was used. The sample collection procedure was approved by the University’s Ethics Committee (Ahvaz Jundishapour University of Medical Sciences). The sample size was calculated using GPower V. 2.9.1.3 software. The most important inclusion criteria for study participants were female patient with primary tumor site on breast, no history of benign breast disease, no chronic inflammatory disorders, and the accessibility of the tissue along with clinicopathologic data of all patients. Female patients previously treated with SAM-interfering drugs, pregnancy, breastfeeding, as well as those missing clinicopathologic data were excluded from the study.
3.2. DNA Extraction and Sodium Bisulfite Treatment of DNA
The DNA was extracted by the phenol-chloroform method and the extraction process was carried out according to the previous study (11). After extraction, the DNA-containing microtube was stored at -20°C.
Afterward, DNA was exposed to sodium bisulfate according to the method previously described (12). In brief, NaOH solution (2 M) was added to 1 - 2 µg of genomic DNA. Denaturation of DNA started efficiently after incubating the mixture for 20 minutes at 37°C.
Then about 30 μL of 10 mM hydroquinone and 520 μL of sodium bisulfite 3 M (PH = 5) were added to the denatured DNA and the surface was coated with a few drops of heavy mineral oil. Next, the solution was incubated at 50°C for 16 hours. Following the incubation, the methylated modified DNA was purified by the DNA purification kit of Roche. Subsequently, 5.5 μL of 3 M sodium hydroxide (NaOH) was added to the modified DNA and incubated at room temperature for 5 minutes. The solution was neutralized by the addition of 100 μL of 5 M ammonium acetate (NH4OAC). Finally, the DNA was precipitated in ethanol.
3.3. Methylation-Specific Polymerase Chain Reaction (MSP-PCR)
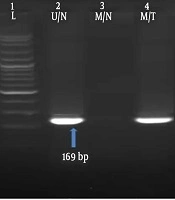
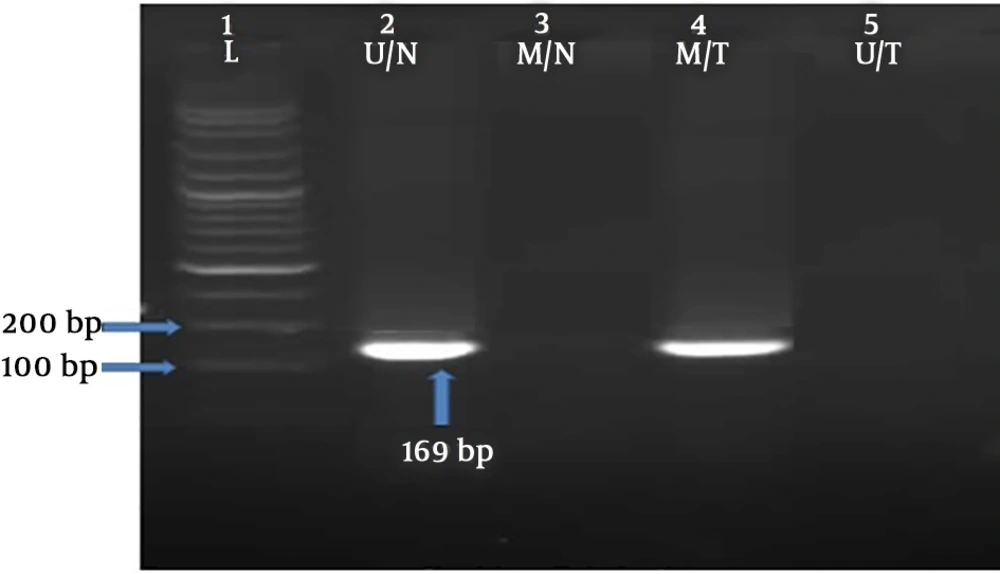
The methylation status of RASSF1A gene was investigated by using methylation-specific polymerase chain reaction (MS-PCR) technique. Two pairs of primers for methylated DNA (M pair) and for unmethylated DNA (U pair) were used that were previously described by Motevalizadeh Ardekani et al. (13), as shown in Table 1. The PCR temperature cycling conditions were as follows: initial denaturation at 95°C for 5 minutes, 40 cycles of 95°C for 30 seconds, annealing at 64°C for 25 seconds, 72°C for 25 seconds, and the final cycle was followed by extension at 72°C for 10 minutes. The PCR products were determined on 1% agarose gel. An image, presenting diverse methylation patterns, was captured (Figure 1).
| Primer | Sequence (5'→3') | Product Size (bp) |
|---|---|---|
| RASSF1A-M | F: GGGTTTTGCGAGAGCGCG | 169 |
| R: GCTAACAAACGCGAACCG | ||
| RASSF1A-U | F: GGTTTTGTGAGAGTGTGTTTAG | 169 |
| R: CACTAACAAACACAAACCAAAC |
3.4. Statistical Analysis
Statistical analysis was performed with SPSS (version 25). P values were estimated using Fisher’s exact test and P < 0.05 was considered statistically significant.
4. Results
4.1. Methylation Analysis of a CpG Island in the Promoter Region of RASSF1A
The methylation status of the RASSF1A promoter was investigated on the DNA samples of 20 sporadic breast cancer tumors (with mean age 48 ± 6.7 years) and 20 adjacent noncancerous tissues of the control groups (with mean age 47 ± 7.6 years). As presented in Table 2, the RASSF1A promoter methylation was not associated with the increased risk or prognosis of breast cancer (P = 0.6).
| RASSF1A Methylation Status | Breast Tumors | Normal Tissues | P Value |
|---|---|---|---|
| MM | 3 (15) | 1 (5) | 0.6 |
| UM | 4 (20) | 5 (25) | 1 |
| UU | 13 (65) | 14 (70) | 1 |
| Total | 20 | 20 |
Abbreviations: MM, fully methylated promoter; UM, semi-methylated promoter; UU, fully unmethylated promoter.
aValues are expressed as No. (%).
The methylated phenotype (MM) was more frequent in breast tumors (3/20 or 15%) compared to the healthy tissues (5/20 or 5%). Also, the semi-methylated phenotype (MU) was detected slightly more commonly in healthy tissues compared to the tumor tissues (25% vs. 20%), but no significant difference was detected between the two groups.
4.2. Association of RASSF1A Methylation with Clinicopathologic Parameters of Breast Cancer Patients
The analysis of the clinicopathologic data of the patients indicated no correlation between RASSF1A promoter methylation and the histopathological grade, familial history of breast cancer, familial history of other cancers, histology. Correlation of ER, PR, and HER2 markers with methylation was observed and RASSF1A methylation was correlated with ER in this locus but had no significant relationship with PR and HER2 (Table 3).
| Parameters | RASSF1A Methylation Status | P Value | |
|---|---|---|---|
| Non-Methylation (N = 17) | Methylation (N = 3) | ||
| Estrogen receptor | 0.04 | ||
| Negative | 1 (5) | 2 (66) | |
| Positive | 16 (95) | 1 (34) | |
| Progesterone receptor | 1 | ||
| Negative | 4 (24) | 1 (34) | |
| Positive | 13 (76) | 2 (66) | |
| Her2 | 1 | ||
| Negative | 10 (59) | 2 (66) | |
| Positive | 7 (41) | 1 (34) | |
| Histopathological grade | 0.7 | ||
| I | 2 (12) | - | |
| II | 7 (41) | 1 (33.3) | |
| III | 6 (35) | 1 (33.3) | |
| IV | 2 (12) | 1 (33.3) | |
| Familial history of breast cancer | 0.5 | ||
| Negative | 14 (82) | 2 (66) | |
| Positive | 3 (18) | 1 (34) | |
| Familial history of other cancers | 1 | ||
| Negative | 5 (30) | 1 (34) | |
| Positive | 12 (70) | 2 (66) | |
| Histology | 0.4 | ||
| Ductal | 15 (88) | 2 (66) | |
| Non-ductal | 2 (12) | 1 (34) | |
aValues are expressed as No. (%).
5. Discussion
One of the most common mechanisms for silencing cancer-related genes is the aberrant methylation of the promoter region, which resulted in downregulation of gene expression. It has been established that the loss of gene expression of important tumor suppressor and growth-regulatory genes that led to cancer can occur via CpG island methylation (14).
While many investigations have shown that CpG island methylation plays a key function in the development of breast cancer, the results were quite variable. The diverse outcomes may be correlated with the use of diverse DNA methylation markers and techniques of analysis. The aim of this study was to compare the DNA methylation pattern in the promoter region of the RASSF1A gene in cancerous and normal tissues. Also, we aimed to verify whether the methylation of RASSF1A gene involved in development of breast cancer correlated with particular clinicopathologic features and hormone receptor expression in patients affected by breast cancer in Khuzestan province.
Epigenetic inactivation of RASSF1A by methylation is identified to be common in carcinomas of lung, ovary, gastric, bladder, nasopharyngeal, and breast (7, 15-18). Based on previous studies, hypermethylation in the promoter region of this gene does not occur in the healthy breast tissue, so the promoter methylation of RASSF1A is a specific tumor phenomenon (6, 19).
The RASSF1A methylation frequency in tumoral and normal tissues was 15% and 5%, respectively (Figure 1), which was lower as compared to previous studies (19-24). Accordingly, our results were higher than the studies by Motevalizadeh Ardekani et al. (13) and Cho et al. (25) showed that the methylation level of RASSF1A was 9.5% and 4%, respectively. Interestingly, no significant difference was revealed in the methylation level of RASSF1A between the cases with breast cancer and the healthy control group.
The association of RASSF1A methylation in patients with breast cancer and healthy controls has been analyzed in several studies (24-33). Burbee et al. (19), Park et al. (24), and Kim et al. (30) detected significantly high frequency of RASSF1A methylation in breast carcinoma than control subjects. In contrast, Cao et al. (26) and Zmetakova et al. (32) revealed no significant difference in methylation frequencies of RASSF1A gene between breast cancer patients and normal controls, which was in line with our results. Furthermore, another study by Brooks et al. demonstrated no significant difference in frequencies of RASSF1A promoter methylation between breast cancer cases and healthy control groups (33). Actually, it is not easy to compare the results between the published data and the present study, where different methods have been used for methylation analysis and diverse CpG sites have been investigated. The exact number of methylated CpG sites as the potential biomarker for breast cancer risk still remains unclear.
We next investigated the correlation between RASSF1A promoter methylation and relevant clinicopathologic parameters including histopathological grade, familial history of breast cancer, familial history of other cancers, estrogen-receptor, progesterone-receptor, Her2 and histology. Our findings revealed that the frequency of RASSF1A methylation was not statistically different at low-grade and high-grade tumors that were in the same line with previous studies (18, 19, 21).
In contrast, we identified a significant correlation between RASSF1A promoter methylation and ER-negative, demonstrating that ER status can have an effect on epigenetic modifications of certain genes. The association of RASSF1A promoter methylation and ER negativity was detected in previous studies (19, 34), while some previous studies were inconsistent with our findings and they found a correlation between ER positivity and RASSF1A promoter methylation (23, 35-38). Also, we demonstrated no significant correlation between RASSF1A promoter methylation in tissue, PR and Her2 markers in our study that is in line with previous studies (20, 37).
Various results in previous studies and the present study can be due to technical problems in the MSP-PCR and DNA extraction from tumor cells, using different methods for evaluation of promoter methylation, different stages of tumor cells and heterozygosity of the alleles of this gene. Also, another limitation of our findings was the small sample size, which included 20 patients and 20 healthy controls. Therefore, large-scale multicenter prospective survey cohorts are required to confirm these findings.
In summary, the status of promoter methylation of the RASSF1A gene in tissue included in our survey was unable to discriminate between breast cancer patients and healthy control groups. More prospective studies should be conducted to assess whether RASSF1A promoter methylation can be an appropriate biomarker for breast cancer early detection.