1. Background
Leishmaniosis is one of the most important parasitic diseases in the world (1, 2). This disease is a complex infection caused by several species of Leishmania genus belonged to family Trypanosomatidae, subfamily Leishmaniinae (3). In the tropical regions, leishmaniosis is considered as the second leading cause of death due to infectious diseases; the overall prevalence is 12 million cases with the annual incidence of 2 to 2.5 million (4).
Currently, leishmaniosis is endemic in 98 countries, but 90% of visceral leishmaniosis (VL) cases occur in the Bangladesh, Brazil, India, Nepal, and Soudan and 90% of cutaneous leishmaniosis (CL) cases occur in Iran, Peru, Saudi Arabia, Brazil, and Syria (4).
According to the reports of the disease control center, annually about 20,000 cases with different types of leishmaniosis occur in Iran. This makes Iran the country with the highest prevalence of cutaneous leishmaniosis in the Middle East (5) and now it is endemic in 15 provinces (6); 2863 CL cases were reported in Central and Southeast regions of Sistan and Baluchistan province, Iran, from April 2008 to March 2014, which indicates the importance of controlling CL in this area. CL is a growing public health problem with several new foci identified in the last years. The endemicity is so high that almost 80% of the rural population contracts the disease before the age of 10 and practically all non-immune newcomers are infected.
Traditionally, in some regions of the word, such as Iran, people are immunized against Leishmania spp. by receiving a specific attenuated vaccine into their hand or foot; the program that is called Leishmanization. Due to the side effects of leishmanization such as the increase of immune system diseases, this method is no longer used (7, 8).
Another approach to immunize against Leishmania spp. is to receive the inactivated or the promastigotes extract vaccines. This method did not have good results either (7). Probably, a combination of lower amounts of antigen and the inactivated vaccine can explain the inefficiency of the vaccine (9). In the next step, researchers used some protein subunits of the parasite as a vaccine that in comparison with the inactivated vaccines showed better results. Nowadays, researchers try to find appropriate proteins and adjutants to improve the performance of this type of vaccines (7). But unfortunately, to date there is no definitive vaccine against the leishmaniosis in the word (10, 11).
Since the vaccination is still one of most effective methods to reduce mortality due to infectious diseases, scientists are looking for a new generation of vaccines to prevent the challenges of leishmaniosis in people (9).
In recent years, many studies are conducted on DNA vaccines as a new vaccination method against leishmaniosis. In comparison with inactivated and subunit vaccines, DNA vaccines can be easily produced and are much cheaper; also, they can activate the T helper type 1 (Th1) CD4 and T-cytotoxic CD8 cells that are significantly effective against the Leishmania species (12-16). Until now, the genes producing different proteins of Leishmania spp. are used to make DNA vaccines and almost good results are obtained. LACK (17-20), GP63 (21-23), PSA2 (24), and ORF are among these proteins (25). Some of the studies showed good results, but some others were not helpful and sometimes the results were contradicted.
As Leishmania spp. are the internal parasites, vaccines that can activate Th1 more than Th2 are more successful. Also, elevated ratio of interferon (IFN)-γ to interleukin (IL)-10 in the body is known as a success factor of vaccination (26). Since DNA vaccines can activate Th1 very well, they can be considered as appropriate vaccines for Leishmania species.
One of the strategies used by researches to increase the efficiency of DNA vaccines is the employment and successive attachment of several effective epitopes simultaneously (recognized by T-cells) and construction of a united polytope. This method showed much better simulation of Th1 (16, 27). The use of polytopes in DNA vaccine is an effective strategy to protect against Leishmania species. In this method epitopes identified by Th1 cells were used to produce more effective DNA vaccines (14, 27).
Since leishmaniosis has high prevalence in Sistan and Baluchistan province, Iran, the current study aimed at investigating the performance of a polytope DNA vaccine constructed by LACK, GP63, and CPC epitopes, and evaluating its efficiency in vivo.
2. Methods
2.1. Plasmid Construction and Purification
The current study used a recombinant pcDNA3.1 expression plasmid consisting of a polytope gene (264 bp) of 3 epitopes of LACK, GP63, and CPC of Leishmania spp. antigens. The plasmid was constructed in the authors’ previous work (28). Vector was transformed into Escherichia coli DH5α, and after plasmid extraction vector was confirmed by polymerase chain reaction (PCR) and digestion enzyme analysis (data not shown). The Qiagen kit was employed to purify plasmid according to the manufacturer’s instructions.
2.2. Immunization and Infectious Challenge
Thirty female Balb/c mice were divided into 3 groups. Group 1 received 100 µL phosphate-buffered saline (PBS) in 3 doses with 2-week intervals intramuscular (IM); the group 2 received pcDNA3 (100 µg in 100 µL) in 3 doses with 2-week intervals (IM) as the first control group; the group 3 received pcDNA3.1/ LAKJB93 (100 µg in 100 µL, eluted in PBS) in 3 doses with 2-week intervals (IM) as the second control group. All animals were challenged subcutaneously (SC) in the right hind footpad with 5 × 105L. major stationary-phase promastigotes (in 50 µL PBS) 2 weeks after the last immunization. Mice were monitored weekly by measuring the footpad swelling, using a metric caliper.
2.3. Blood Collection and Serum Isolation
Blood samples were collected twice, 2 and 6 weeks after the last immunization. Sera were isolated from whole blood samples and stored at -20°C until use.
2.4. Cytokines Assay
Spleen of immunized animals was harvested 6 weeks after challenge and spleen cells were collected by conventional procedures, and then, the red blood cells were removed by centrifugation over Ficoll-Hypaque. Spleen suspensions (2 × 106 per well) were stimulated with 5 µg/mL soluble Leishmania antigen (SLA) in RPMI-1640, supplemented with 10% heat-inactivated fetal bovine serum, 50 µM β-mercaptoethanol, and 40 µg/mL gentamycin. Culture supernatants were collected after 36 or 72 hours to determine IL-4 and INF-γ, respectively. Levels of INF-γ and IL-4 were determined by the sandwich enzyme-linked immunosorbent assay (ELISA) technique using R&D system kits.
2.5. Antigen Preparation
To prepare SLA, after a few passages in liquid culture, late-log-phase promastigotes were collected (29). Briefly, after washing 2 × 108 promastigotes/mL in 5 mL of cold PBS 3 times, the promastigotes were frozen and thawed for 5 times. The suspension was centrifuged at 8000 g for 20 minutes at 4°C. Supernatants containing SLA were collected and stored at -70°C. The protein concentrations were estimated by the Bradford method.
2.6. Evaluation of Humoral Immunity
Pooled sera were prepared from each group of mice 2 weeks after the last immunization, and 6 weeks after the challenge. Production of IgG1 and IgG2a antigen specific antibodies was measured against LAKJB93.
Briefly, 96-well plates were coated with recombinant proteins (500 ng/well) and incubated at 4°C overnight. They were washed 3 times with PBS plus 0.05% Tween 20, plates were blocked with 1% bovine serum albumin (BSA) in PBS for 2 hours at 37°C; then, 100 μL of pooled sera were added and incubated for another 2 hours at 37°C (1:50 dilution). Anti-mouse IgG1 (1:10000, Zymed, Burlington, ON, Canada) and IgG2a (1:500, southern Biotech Birmingham, USA) conjugated with Biotin were added. After 2 hours at 37°C, streptavidin-horseradish peroxidase (BRL, Gaithersburg, MD) was added (1:1000). After incubation for 1 hour at 37°C, Conjugates were visualized with O-phenylenediamine (OPD), the reaction was stopped with H2SO4 (4 N), and the absorbance was read at 492 nm.
2.7. Statistical Analysis
All statistical analyses were performed with SPSS version 19. One-way analysis of variance (ANOVA) and the post-hoc Tukey test were used to differentiate between the analyzed groups. All results were expressed as mean ± SD. For all tests, a P value of < 0.05 was considered as statistically significant.
3. Results
3.1. Detection of Systemic Immune Response
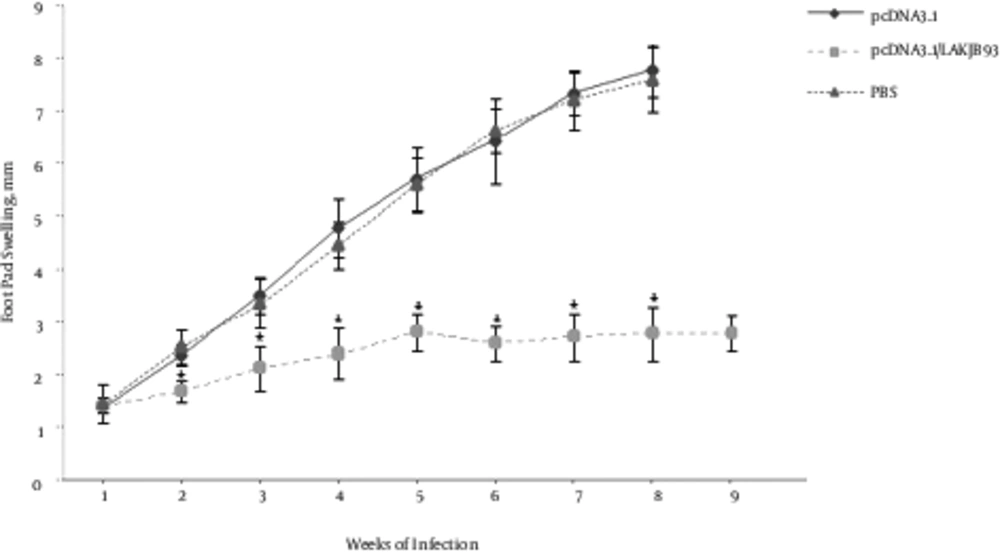
To determine the efficiency of the DNA vaccination, mice were infected with L. major 2 weeks after the last vaccination and footpad lesion sizes were measured 9 weeks after the infection. As shown in Figure 1, mice immunized with DNA vaccine, compared with the control group cases, had significantly smaller lesion sizes (P < 0.05). To assess the efficiency of DNA vaccine in the induction systemic immunity, all mice were infected in the opposite footpad to which they received DNA vaccination.
Infectious Challenge of Balb/c Mice Against L. major, Immunized with DNA Vaccine Encoding Fusion Antigen. Mice (5 per group) were immunized with 100 µg pCDNA3.1/LAKJB93, or pCDNA3.1, or equal volume of PBS as test, the sham, and control groups, respectively. Two weeks after the last immunization, mice were challenged by the inoculation of 5 × 105 amastigote forms of L. major in the right footpad. Lesion sizes were monitored using a Vernier caliper and calculated by subtracting the size of contralateral-uninfected footpad. No significant differences were observed between pCDNA3.1 and PBS groups. However, lesion sizes significantly decreased in pCDNA3.1/LAKJB93 group, compared with the control groups (pCDNA3.1 and PBS) after the week 2 (*P < 0.05). ANOVA and post-hoc Tukey test were employed. Data are expressed as mean ± SD of lesion sizes (in millimeters) from 5 mice per group.
3.2. Measurement of IFN-γ and IL-4 Production in Infected Mice
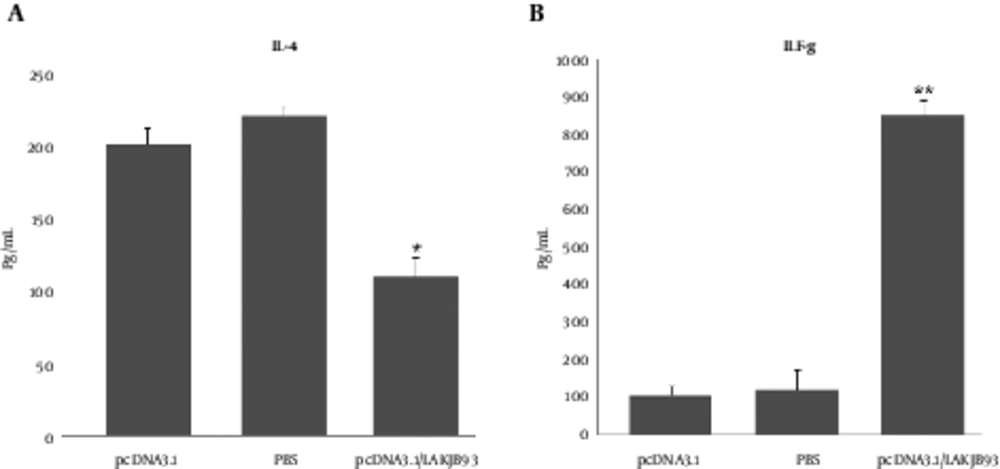
Production of IFN-γ and IL-4 from spleen cells was assessed 6 weeks after the infection challenge. As shown in Figure 2, spleen cells from mice vaccinated with pCDNA3.1/LAKJB93 significantly developed more IFN-γ and IL-4, compared with the control group (pCDNA3.1 or PBS) (P < 0.05).
Production of IL-4 (A) and IFN-γ (B) from Spleen Cells of Vaccinated Balb/c Mice Infected with L. major 6 Weeks After the Infection. Mice in each group (n = 5) were euthanized and the spleens were harvested 6 weeks after the infection. Single-cell preparations plated in triplicate in 96-well micro titer plates; 48 hours later, supernatants were harvested and ELISA measured IFN-γ and IL-4 content. Significant differences were observed between pcDNA3.1/LAKJB93 and the control groups (pCDNA3.1 and PBS) (*/**P < 0.05) for the amounts of IL-4 and IFN-γ. ANOVA and the post-hoc Tukey test were used for all analyses.
3.3. Measurement of Specific IgG1 and IgG2a in Vaccinated Mice
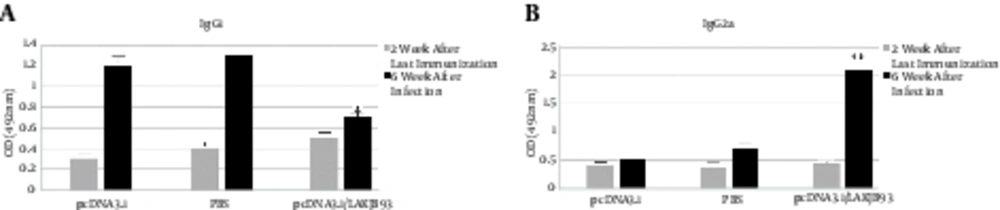
Production of specific IgG1 and IgG2a antibodies was measured to provide an in vivo assessment of the pattern of cytokine production. As shown in Figure 2, mice that were vaccinated with pCDNA3.1/LAKJB93 and developed an effective immune response had substantially higher levels of specific IgG1 and IgG2a antibody titers, compared with the control groups (pCDNA3.1 and PBS) (P < 0.05).
Sera (10 mice per group) were collected 2 weeks after the last immunization and 6 weeks after infection from mice vaccinated with pCDNA3.1/LAKJB93 and control groups (pCDNA3.1 and PBS); 1:100 dilutions of sera were tested for the specific IgG2a and IgG1 fusion genes. IgG2a and IgG1expressions significantly increased in pCDNA3.1/LAKJB93 group, compared with the control groups (*/**P < 0.05). Data are expressed as mean ± SD. ANOVA and the post-hoc Tukey test were used for all analyses.
4. Discussion
Based on the obtained results of the current study, the DNA vaccine made from 3 epitopes of LACK, GP63, and CPC genes (pcDNA3.1/LAKJB93) of Leishmania spp. could immunize the Balb/c mice against leishmaniosis.
The DNA vaccines are a construct of nucleic acids that nowadays are in focus of attention due to their low price and high efficiency (30). These types of vaccines, in comparison with inactive vaccines and subunit vaccines, have more ability to activate Th1 CD4 and T-cytotoxic CD8 cells and longer immunity responses (12-16, 31).
As Leishmania spp. are intercellular parasites, a vaccine that can activate Th1 more than Th2 is more successful; also, the high ratio of IFN-γ to IL-10 is a promising sign for the success of vaccination (26). Long protection against Leishmania spp. is another parameter of an ideal vaccine, which can be obtained by creating memory T-cells (probably both CD4 and CD8 cells) (17, 32, 33). All of these factors can be achieved by a well-designed DNA vaccine (12, 33, 34).
Since determination of Th1 and Th2 patterns is a main in vivo evaluation, the current study measured IL-4 and IFN-γ levels. Results of the current study showed that polytope DNA vaccine can stimulate Th1 due to the increase of IFN-γ level.
Ahmed et al., used 6 different genes of Leishmania spp. as candidates of DNA vaccine; these genes included GP63, PSAS.P20, LeIF, ribosomal like and the different parts of LACK gene. Each of these DNA vaccines could not completely protect the mice against L. major. However, among these DNA vaccines, the LACKP24 had the highest efficiency; LACKP24 included LACK gene nucleotides of 465 to 939; the current study also used this gene as well as the 2 other genes as a DNA vaccine. In a study conducted by Ahmed et al., the effect of DNA vaccines was investigated individually; it was proposed that if they were used simultaneously in the form of a DNA vaccine, better immunity would be obtained (7).
Dominguez-Bernal et al., made a DNA vaccine that expressed the 6 genes of Leishmania spp. simultaneously in the form of a polyprotein including A2, H4, H3, H2B, H2A, and Hsp70. This vaccine had high efficiency and could protect the mice against the CL caused by L. major. This vaccine led to the increase of producing IFN-γ, IL-17, and NO 7 weeks after the injection of PI; it also reduced the IL-4 production and consequently led to the increase of Th1 activity (31); the current study also showed that polytope DNA vaccine can be used as a vaccine that protected mice from L. major infection.
Doroud et al., delivered a cocktail DNA vaccine encoding cysteine proteinases type I, II, and III with solid lipid nanoparticles potentiating protective immunity against L. major infection; results of their research showed that cocktail DNA can protect mice (15), although nanoparticles were not used for delivery, it was proved that polytope DNA vaccine could be useful.
The current study also used a combination of multiple epitopes to elevate the performance of a DNA vaccine against L. major, a combination of the 3 epitopes of LACK, GP63, and CPC genes of L. major, which were simultaneously used in a modified pCDNA.3 vector as a DNA vaccine. In vivo studies indicated that the levels of IgG1 and IgG2a significantly increased in vaccinated mice (P > 0.05). This result was combined with low-level IL-4, but high IFN productions (P > 0.05) showed that the vaccination was successful. In addition, the challenge test conducted on 5 mice in each group revealed that the vaccine could lead to long-time resistance against L. major infection (P > 0.05) (infected mice were monitored 9 weeks after the infection).
4.1. Conclusion
The employed DNA vaccine could stimulate the cellular immunity and protect the mice against leishmaniosis; thus, this vaccine can control the health problem and increase the public health in Iran.