1. Background
Hepatocellular carcinoma (HCC) is the most prevalent primary liver malignancy in adults and is the third main cause of cancer-related deaths in humans. Viral infections, including hepatitis B virus (HBV) and hepatitis C virus (HCV), are the leading causes of HCC (1). The underlying mechanisms of HCC include a complex of multistep molecular signaling pathways that handle alterations in the genetic figures of the virus and/or the host (2). Nevertheless, the precise molecular mechanisms by which HBV contributes to the development of HCC are not yet completely understood. However, recent studies have identified that some biological pathways are more involved in the underlying mechanisms of HCC development, including the wingless-related integration site (Wnt) pathway, EGF-R, Ras/MAPK, and PI3K/Akt (3).
The Wnt pathway is a highly conserved critical pathway that is important in the regulation of cell polarity and differentiation and plays a role in several tumors such as HCC (4, 5). β-catenin is the key element of the Wnt pathway that may be mutated and/or overexpressed in an aberrant state of this pathway (6-8). In most adult tissues, the Wnt pathway remains inactive or steady. This physiologic state is regulated through phosphorylation of β-catenin by the Gsk3β and CK enzymes that lead to degrading β-catenin, thus preventing its transportation to the nucleus (9). However, in an active Wnt state, the Wnt ligands interact with the Frizzled class receptors (FZD) receptors, thereby stabilizing cytoplasmic β-catenin, leading to nuclear transportation, which, in turn, induces transcription through TCF/LEF and target genes involved in cell proliferation (10).
Hepatitis B virus is a major leading cause of HCC, with 887,000 deaths in 2015, largely due to cirrhosis and HCC (11). Moreover, HBV-related HCC has shown an increasing trend in the last decades in developed countries (12). Although the involvement of HBV-related mechanisms in the development of HCC is not yet completely understood, some features of HBV are proposed, including higher viral loads in chronic HBV infection (13), HBV genotypes C and F (14), integration of viral genes into the human genome, viral proteins and last but not least, mutations in viral proteins (HBx, HBsAg) (13-15). As a pleiotropic protein, HBx is known for the transcriptional regulation and activation of a variety of host genes involved in epigenetic modification and control of cell proliferation. Therefore, the investigation of its oncogenic potential has been hyped recently as an attractive field of interest (16). Recent studies demonstrated that HBx has potential roles in the regulation of a variety of signaling transduction pathways, including Wnt/β-catenin, Ras/MAPK, SAPK/JNK, and NF-κB pathways (17, 18). Several lines of studies indicated that HBV also produces mutant forms of HBx that are involved in carcinogenesis through the epigenetic regulation of several pathways and/or tumor suppressor genes (19, 20).
2. Objectives
In this work, we attempted to study the Wnt pathway with a comprehensive gene expression analysis along with the virological characteristics for the elucidation of potential interactions between HBV and Wnt pathway status.
3. Methods
3.1. Patients and Samples
A cross-sectional study was conducted using fresh frozen (FF) samples taken of surgical resection by fine needle biopsy in hospitals affiliated to Iran University of Medical Sciences (IUMS). The FF samples included HCC tissues and some other tissues that were later identified as histologically normal. The study retrospectively investigated formalin-fixed paraffin-embedded (FFPE) samples, including HCC and non-tumor tissues, as well. Indeed, the control group included liver samples routinely gathered for medical reasons and later determined as histologically normal. The stored samples had been collected from March 2010 to August 2019, and FF samples from May 2017 to September 2019. The FF and FFPE samples were histologically examined by an expert pathologist for the confirmation of HCC or normal histology. The serologic data including the level of liver enzymes, HBV, and HCV seromarkers, were obtained from the medical records of patients in the hospitals. All participants signed informed consent forms, and the proposal of this study was approved by the Research Ethics Committee of IUMS (ethics code: IR.IUMS.FMD.REC 1396.9321540004).
3.2. DNA Extraction
One milligram (mg) of each tissue sample was subjected to DNA isolation using DNA extraction kits (MN, Macherey-Nagel GmbH & Co. KG) according to kit instructions with minor modifications. The FFPE samples were first cut in 15-µm thicknesses, followed by paraffin removal using xylene and washing with 100% ethanol. Subsequently, 1 mg of the obtained tissue was used for DNA extraction, as mentioned above. The concentration and purity of isolated DNA were checked by BioPhotometer and agarose gel electrophoresis, and DNA isolation was confirmed by performing a PCR for β-globin, as described previously (21).
3.3. RNA Extraction and cDNA Synthesis
A tissue RNA extraction kit (NucleoSpin® total RNA, MN, Macherey-Nagel GmbH & Co. KG) was used for the extraction of RNA from FF samples. The FFPE samples were first cut in 15 µm thicknesses, followed by paraffin removal using xylene and washing with 100% ethanol. For the improvement of RNA isolation from FFPE samples, we used a further processing method, and then the processed samples were subjected to the above-mentioned kit, as described in the Supplementary section. The purity and concentration of obtained RNA were checked by a BioPhotometer (ratio of 260/280 was optimized between 1.7 and 2). The concentration and integrity of RNA were observed with agarose gel electrophoresis.
For cDNA synthesis, extracted RNA was treated with DNase I (Yekta Tajhiz Azma, YTA, Iran) to remove any contaminating genomic DNA. Next, the cDNA synthesis was done by using random hexamer primers according to the instruction of the cDNA synthesis kit (Fermentas, Thermo Fisher Scientific TR Limited, Waltham, Massachusetts, United States).
3.4. Determination of HBx Mutations
The HBV infection was investigated by partial amplification of the small S gene, as described previously (22-24). A conventional nested-PCR was performed to amplify the HBx gene, as described previously, with some modifications (24). For the determination of HBx mutations, PCR products were sequenced and aligned with an attributed reference sequence in BioEdit software (GQ183486).
3.5. Assessment of Intrahepatic Viral Load
Five microliters (µL) of the extracted DNA were used for the determination of the viral load in liver tissues according to a previously described quantitative method with minor modifications (23). The primers and probe sequences were as follows: BSF: 5’-TCGTGGTGGACTTCTCTCAA-3’, BSR 5’-AGGACAAACGGGCAACATAC-3’, probe: 5’-FAM-AACTACCGTGTGTCTTGGCC-TAMRA-3’. These primers correspond to a highly conserved region of the S gene that covers all HBV genotypes. The reaction was performed in a three-step program: Initial denaturation at 95°C for 5 min, followed by 45 cycles at 95°C for 15 s, 58°C for 15 s, and 72°C for 20 s. A cloned segment of the small S gene was serially diluted tenfold and used for the construction of standard curve and quantification in each run. The concentration of intrahepatic HBV load in the samples was expressed as copies/µL of extracted DNA (for all samples, 1 mg of tissue was used for DNA extraction and eluted from the spin filter with 50 µL elution buffer).
3.6. Investigating the Expression Level of Wnt Pathway Genes
Among the key elements of the Wnt pathway, 14 genes were selected for the assessment of gene expression using real-time PCR. The selection of these genes was based on a literature review of previous in-vitro and/or in-vivo studies that demonstrated the deregulation of different Wnt genes (primers designed and used for the gene expression assay are provided in the supplementary method). Due to the baseline degradation of RNA in FFPE samples ( Supplementary File), there was a minor shift in the raw Ct data between paired FF and FFPE samples. Although there was a substantial correlation between the Cts of paired FF-FFPE samples, the mean Pearson’s correlation coefficient was 0.86 ± 0.11. The amplification program was carried out in a Rotor-Gene 3000 instrument using SYBR-PCR master 2X (Yekta Tajhiz Azma, YTA, Iran). Samples were tested in duplicate, and the mean cycles of threshold were considered. Values were normalized relative to the expression of two internal housekeeping genes (glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and hydroxymethylbilane synthase (HMBS)). The 2-ΔΔCt formula was used for the calculation of relative gene expression levels. In this way, ΔCt is equal to the Ct of target gene minus the mean Ct of GAPDH and HMBS, and ΔΔCt is equal to tumor tissue ΔCt minus normal sample ΔCt (25).
3.7. Methylation Analysis
Downregulated genes were analyzed further by methylation specific-PCR (MSP). In this method, 2 µg of extracted DNA underwent bisulfite genomic treatment (MaxSpin Bisulfite Modification Genomic DNA, Maxcell Company, Iran). The MSP analysis was applied for SFRP1, SFRP5, and CDH1, as described previously (18, 26). The hypermethylation and suppressed status of SFRP1 had already been established among different cancer cell lines such as HeLa and Caco-2 (27, 28); thus, we used HeLa and Caco-2 cells as positive controls of methylated DNA.
3.8. Statistical Analysis
All qualitative and quantitative variables were analyzed by the chi-square (χ2) test or Fisher’s exact tests, Mann-Whitney test, and student t-test using SPSS version 25. The measurement of ΔΔCt and folding of expression levels was conducted in Excel. The graphical analysis of gene expression was performed by Graph Pad Prism 6.
4. Results
4.1. Baseline Characteristics of the Population
A totally of 161 samples were examined including HBV-HCC (n = 79), non-viral HCC (n = 41), and tissues with normal histology (n = 41). Demographic and clinical data of the patients are summarized in Table 1. The mean ALT enzyme level was 66.6 ± 43.2 IU/L, and the mean AST level was 96.4 ± 70.1 in the tumor group that were significantly higher than the normal range (P = 0.009 and P = 0.005, respectively). In the control group, samples with normal results in terms of liver enzymes, blood sugar, LDL, and bilirubin levels were selected for the gene expression assay. Besides participants of the control group of gene expression analysis, all had a negative result for viral markers.
| Characteristics | HBV-HCC (N = 79) | Non-Viral HCC (N = 41) | Normal Liver (N = 41) |
|---|---|---|---|
| Gender | |||
| Male | 61 (77.2) | 28 (68.3) | 22 (62.8) |
| Female | 18 (22.8) | 13 (31.7) | 13 (37.1) |
| Age | |||
| Mean ± SD | 60.2 ± 12.4 | 59.2 ± 14.2 | 50.5 ± 14.5 |
| < 40 | 4 (5.1) | 5 (12.2) | 7 (20) |
| 40 - 50 | 9 (11.4) | 5 (12.2) | 9 (25.7) |
| 51 - 60 | 23 (29.1) | 6 (14.6) | 11 (31.4)2 |
| > 60 | 43 (54.4) | 25 (61) | 8 (22.8) |
| HBV+ | 79 (100) | 0 | 16 (39) |
| Cirrhosis | 23 (29.1) | 7 (17.1) | - |
aValues are expressed as Mean ± SD or No. (%).
4.2. Viral Characteristics
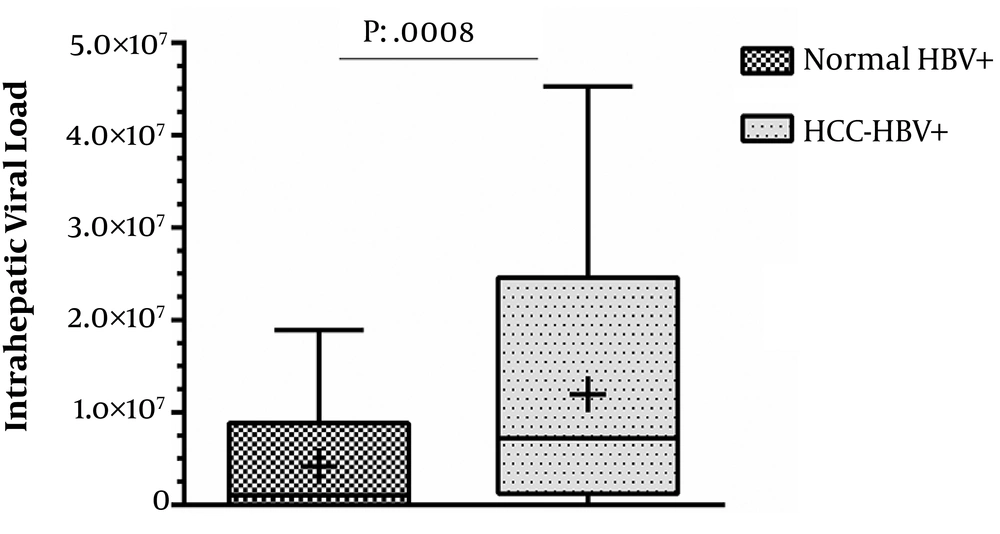
Tissue samples were subjected to DNA isolation and then tested for HBV-DNA. Overall, 59% of the samples were found to be positive for HBV, while the rate was 79/120 (65.8%) among tumor samples. All the HBV-positive tissues were investigated for the determination of intrahepatic viral load. Among 79 HBV-positive tumor samples, 14 (17.7%) had no detectable liver viral load based on real-time PCR. The mean intra-hepatic viral load among HCC cases was 12 × 106 ± 11 × 106 copies/µL of extracted DNA while it was 4.2 × 106 ± 5.6 × 106 copies/µL among HBV-positive tissues with normal histology (P = 0.0008, Figure 1). None of the participants had a positive co-infection result for seromarkers of HCV, HDV, and HIV.
4.3. Expression Level of Wnt Pathway Key Elements
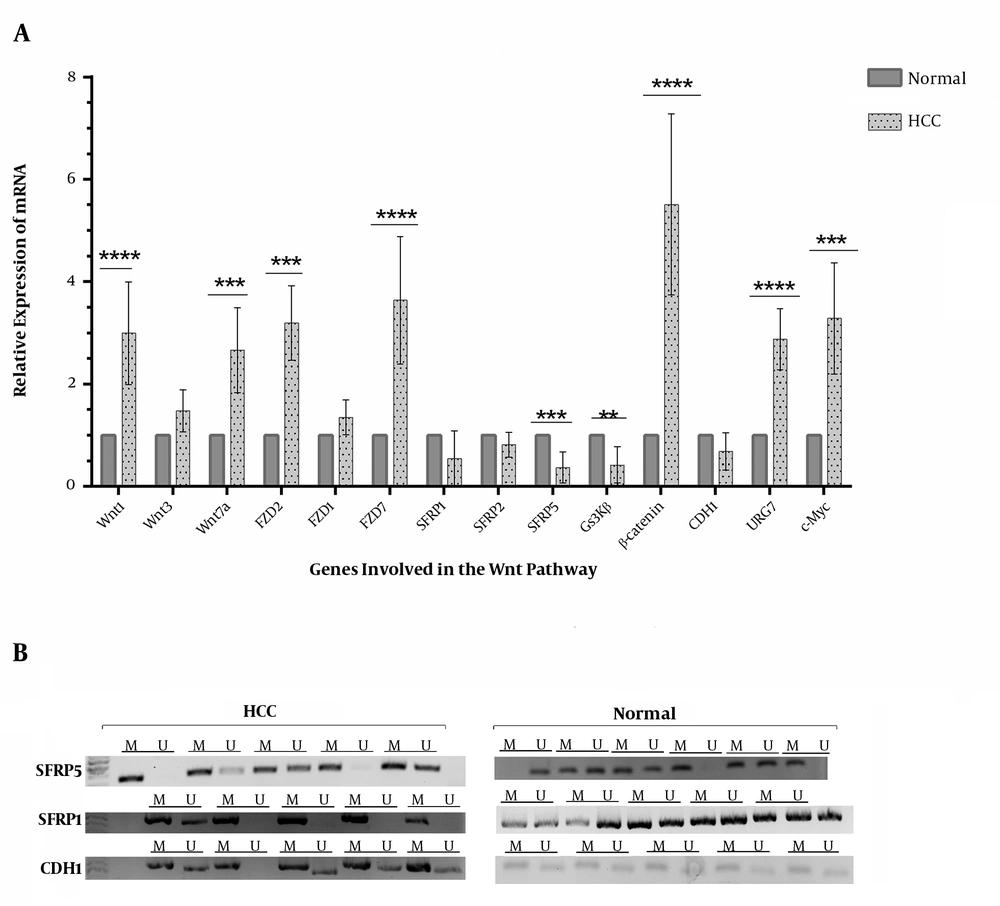
The results of real-time PCR showed an aberrant expression pattern in genes that were assessed in the Wnt pathway (Figure 2). The results showed an increased expression level for Wnt-1, Wnt-7a, FZD2, FZD7, β-catenin, URG7, and c-Myc among the HCC cases. Although there was a significantly decreased expression level for SFRP5 and GSK3β, the observed decreases for CDH1 and SFRP1 were not significant (Figure 2). This result was coherent with the result of MSP analysis, which showed the hypermethylation of SFRP5, SFRP1, and CDH1 genes (Figure 2).
Relative mRNA expression of genes involved in the Wnt pathway among controls (n = 25) and HCC tumors (n = 91); A, differential expression of Wnt genes among two groups (the results were also resolved and semi-quantified on the agarose gel; data are shown in the Supplementary File); B, the MSP assay for CDH1, SFRP1, and SFRP5 that showed an increased methylation level and thus a low expression level; the intensity of the bands was analyzed by GelCuant software (M, methylated DNA; U, unmethylated DNA). *, P < 0.00005.
4.4. Expression Level of Wnt Genes in Association with HBV
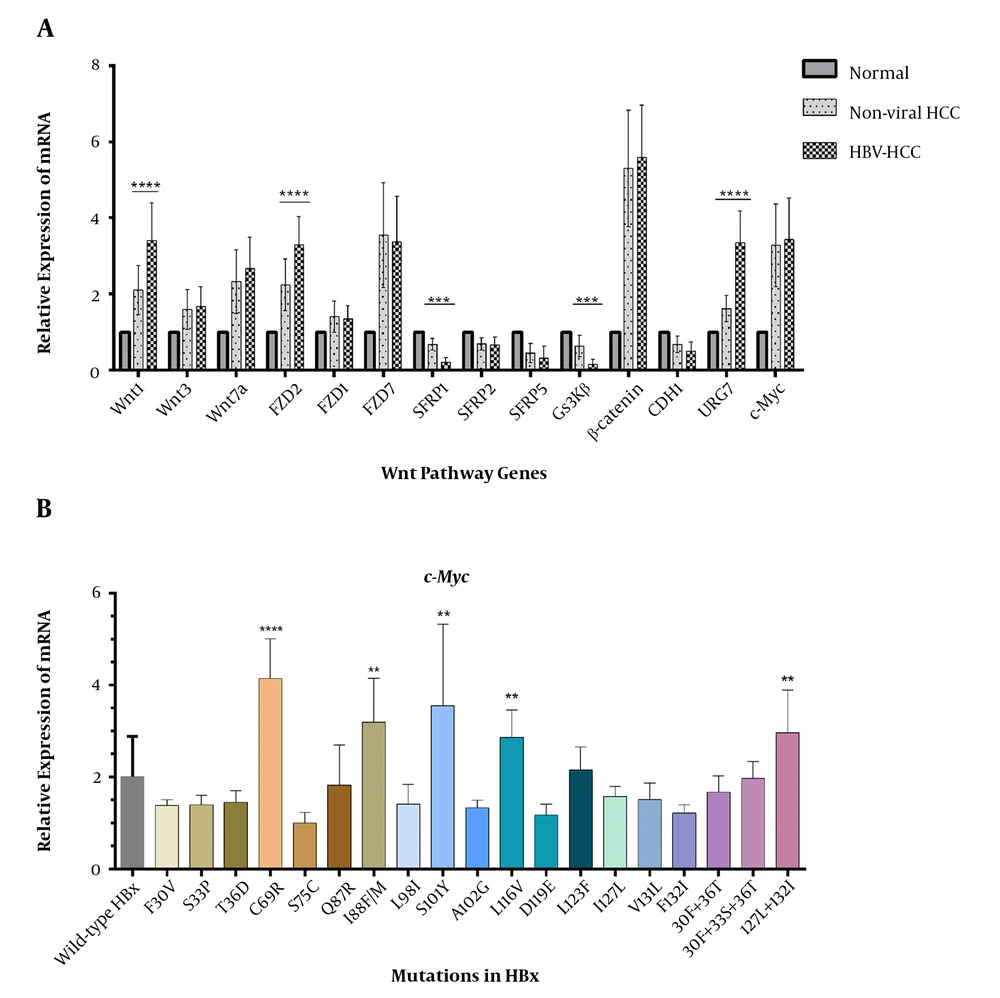
The association of HBV infection with the deregulation of Wnt genes was investigated. There was a more aberrant expression level among several studied genes in HBV-HCCs than in non-viral ones (Figure 3). The expression levels of Wnt-1, FZD2, and URG7 were much higher in HBV-infected HCC tumors, and there was a significant down-expression for SFRP1 and GSK3β (Figure 3).
4.5. Correlation Between the Level of β-catenin and c-Myc
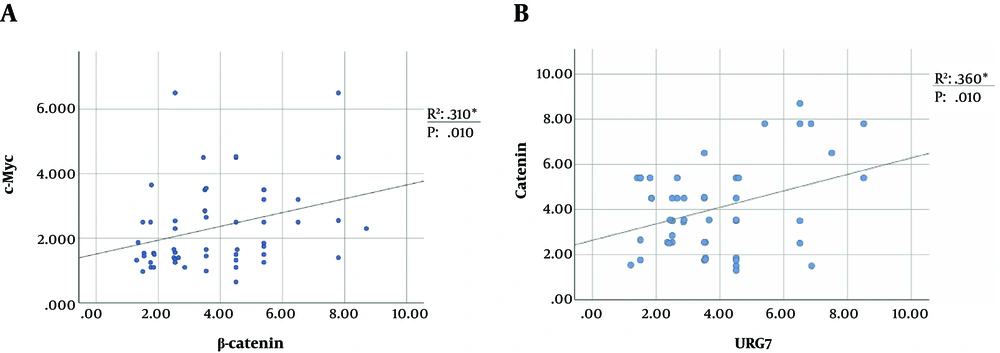
The higher expression of β-catenin leads to cytoplasmic accumulation and further nuclear translocation of this protein, which induces TCF/LEF-mediated transcription of targeted genes. Downstream target genes of the pathway include c-Myc, Cyclin D1, CCN2, connective tissue growth factor (CTGF), (CCN5) Wnt1 inducible signaling pathway protein subfamily 2 (WISP2), c-jun, and c-fus, whose upregulation causes transformation and cancer progression in various tumors (5, 9, 29, 30). The cellular proto-oncogene c-Myc is widely accepted as an upregulated tumor factor in liver malignancies (31, 32). Therefore, we assessed the expression level of c-Myc as one of the downstream targets of the Wnt/β-catenin pathway. Tumor samples with higher expression of β-catenin were significantly correlated with the higher level of c-Myc expression (Figure 4). Therefore, the evaluation of the c-Myc level can be used as a reliable consequence of Wnt pathway activation, as indicated previously (31, 32).
4.6. Wnt Pathway in Association with HBx Mutants
To check the potential effect of different HBx mutants on the Wnt pathway, the HBx region of the HBV genome was sequenced in both HCC-HBV and normal groups. The results revealed that F30V, T36D, I88F/M, and F132I were the most prevalent HBx mutations observed in HBV-HCCs (Table 2). The prevalence of F30V, T36D, I88F/M, and I127T/L mutations was much higher in tumor samples than in normal HBV cases, among which I127T/L was statistically significant.
| Mutation | Normal HBV+ (N = 16) | HCC-HBV (N = 46) | P | Function | Ref. |
|---|---|---|---|---|---|
| F30V | 5 (31.2) | 24 (52.2) | 0.060 | Anti-apoptotic for HBx, related to HCC, reduction of HBV replication | (19) |
| S33P | 3 (18.7) | 9 (19.5) | 0.302 | - | - |
| T36D | 3 (18.7) | 28 (60.9) | 0.050 | - | - |
| C69R | 5 (31.2) | 7 (15.2) | 0.203 | - | (33) |
| S75C | 3 (18.7) | 9 (19.5) | 0.302 | - | - |
| Q87R | 4 (25) | 4 (8.7) | 0.164 | - | (34) |
| I88F/M | 3 (18.7) | 27 (58.7) | 0.051 | - | (20) |
| L98I | 4 (25) | 12 (26.1) | 0.126 | Binding to DDB1 | (35) |
| S101Y | 3 (18.7) | 13 (28.2) | 0.228 | Core promoter | (36, 37) |
| A102G/V | 7 (43.7) | 15 (32.6) | 0.422 | - | - |
| L116V | 3 (18.7) | 8 (17.4) | 0.511 | Core promoter, postoperative survival in HCC | (36) |
| D119E | 5 (31.2) | 14 (30.4) | 0.453 | Core promoter | (37) |
| L123F | 5 (31.2) | 6 (13) | 0.246 | BH3-like motif, Core promoter, EnhII, NRE | (31, 36) |
| I127L | 1 (6.25) | 16 (34.8) | 0.043 | Progression to HCC | (20) |
| V131L | 9 (56.2) | 18 (39.1) | 0.531 | - | (20) |
| F132I | 6 (37.2) | 21 (45.6) | 0.249 | - | (20) |
| Double 30F - 36T | 1 (6.25) | 9 (19.5) | 0.221 | - | - |
| Triple 30F - 33S - 36T | 0 | 3 (6.5) | 0.258 | - | - |
| Double 127L + 132I | 2 (12.5) | 6 (13) | 0.292 | - | - |
aValues are expressed as No. (%).
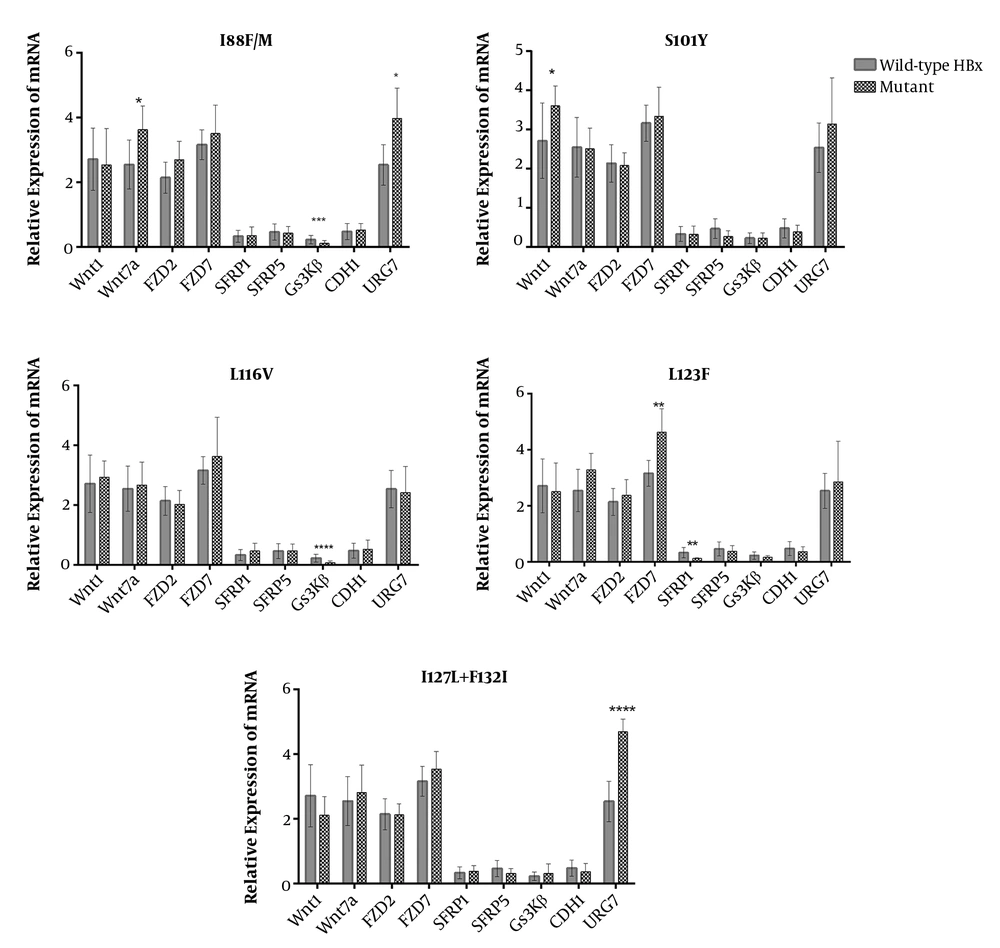
We also surveyed which mutations were more likely associated with deregulations that were in association with HBV-HCCs (deregulated genes found in Figure 3). Comparative analysis between the expression levels of c-Myc among samples harboring different HBx mutants revealed that C69R, I88F/M, S101Y, L116V, and a double mutation at I127 and F132 had roles in the aberration of the β-catenin pathway (Figure 3B). Then, the deregulated genes were analyzed in terms of mRNA expression among cases with different HBx mutations. It was determined that mutations at the protein binding domain of HBx (aa 51 - 151) had a significant impact on deregulation. Overall, URG7 and glycogen synthase kinase 3 beta (GSK3β) were the most prominent genes whose expression was affected by HBx. Mutations, including I88F/M and L116V, were independently associated with the significant depression of GSK3β. There was also a higher expression of Wnt1 among samples with S101Y mutation. The double mutation I127L + F132I was associated with the overexpression of URG7 (Figure 5). The significance of all mutations of HBx in association with the Wnt genes is illustrated in the Supplementary section (Supplementary File).
5. Discussion
Chronic HBV infection is now determined as a principal risk factor responsible for the development of HCC. However, the molecular mechanisms of HBV involvement are not yet clarified. This may be due to different etiologies and several risk factors contributing to HCC, such as aflatoxin, alcohol, other viruses, heterogeneity, and geographical distribution of patients. Meanwhile, the deregulation of some critical pathways, such as the Wnt pathway, is now known as distinctive in the development of HCC. Therefore, the gene expression profile of the Wnt pathway and HBV characteristics were investigated in this research.
In this study, the profiling of mRNA expression was applied for Wnt pathway genes and downstream targets. There was an aberrant Wnt pathway among HCC tumors. We also observed the overexpression of Wnt-1, Wnt-7a, FZD2, URG7, and β-catenin, and down-regulation of SFRP5 and GS3Kβ. The deregulation of different Wnt pathway genes has been repeatedly described in the recent decade (1, 31, 32), which supports the result of the current study.
Mutations in the CTNNB1 gene are major activators of the Wnt pathway with an elevated figure of cytoplasmic β-catenin accumulation. Tumor samples of this study had been previously surveyed for CTNNB1 mutations; cases with mutations in the hot spots of β-catenin had elevated levels of β-catenin and had correlations with the increased expression of cell-myelocytomatosis (c-Myc) (38). Nevertheless, previous studies showed nuclear accumulation of β-catenin in the absence of activating mutations in hot spots of the CTNNB1 gene (39, 40). Therefore, there might be other external or internal factors disrupting the pathway and contributing to the degrading complex (adenomatous polyposis coli (APC), protein phosphatase, component of the beta-catenin destruction complex required for regulating CTNNB1 levels (Axin), GSK3β, and casein kinase 1 family (CK1)) and cytoplasmic stabilization of β-catenin. HBV is a potential external factor in modulation of the Wnt pathway with the benefit of having proteins with some protein binding activities such as HBx, S, and/or Core proteins, which can dysregulate the canonical Wnt pathway (14, 16, 18, 19).
There is a consensus that a higher viral load of HBV is correlated with a greater risk of exacerbation of liver diseases such as liver cirrhosis (LC) and HCC (14, 41, 42). Generally, active HBV infection is measured by serum viral loads for therapeutic objectives; however, hepatocytes are the main replicative targets of HBV, and the liver is the most important tissue for identifying the course of HBV infection (43). Our results showed higher intrahepatic viral loads in the tumor group than in the non-tumor group, as described in some previous studies (43-45). Moreover, previous studies found a direct correlation between serum and hepatic viral load (44, 45).
The activation of the Wnt pathway causes the activation and overexpression of downstream target genes such as c-Myc that can be assessed for the determination of Wnt pathway status. In the current study, tumor samples with elevated levels of β-catenin were significantly correlated with the increased expression of c-Myc; this finding is in line with previous reports (31, 32, 46). Thus, the surveillance of c-Myc can be used as an authentic consequence of active Wnt pathway and overexpression of β-catenin, as described previously (31, 32). However, the result of the current study is based on the expression level of mRNA and needs further assays with the assessment of protein expression levels.
Moreover, it has been found that HBx is a binding partner of APC that can displace β-catenin from its degradation complex (16), thus elevating the cytoplasmic accumulation of β-catenin and subsequently leading to the overexpression of c-Myc. In this study, F30V was the most prevalent HBx mutation; however, it has been previously described concerning HCC and reduced replication efficiency of HBV (19). Gene expression analysis determined that tumor samples with I88F/M and L116V were suppressed in the level of GSK3β, and the double mutation of I127L + F132I was associated with the overexpression of URG7. Moreover, URG7 is a recently discovered spliced variant of the ABCC6 gene, which is upregulated by the induction of HBx (47). This protein has been shown to activate the β-catenin pathway through either inhibiting GSK3β or transactivating the promoter of the CTNNB1 gene (10). Besides, in this study, there was a positive correlation between the mRNA level of URG7 and β-catenin.
The basal Core promoter (BCP) region of HBV harbors most mutations that highly increase the risk of HCC (14, 48). It is noteworthy to mention that most of the regulatory elements of HBV such as enhancer II, Core promoter, BCP, and direct repeat 1 (DR1) are completely or partly within the HBx coding region (48). Therefore, mutations in HBx can affect the HCC progression through the regulation of HBV replication and/or via direct interaction of HBx with host regulatory proteins, as discussed above. In the BCP region, there were some mutations among which, I127T/L was most prevalent; it is a well-known HBx mutation related to a higher risk of HCC (20). This mutation, along with F132I, was associated with the overexpression of URG7.
The protein binding capability of HBx has been shown to target DNA methyltransferases and subsequently downregulate the host genes (48, 49). This phenomenon has been identified for HBx-mediated suppression of CDH1 and SFRP1 genes (18, 26, 50). Furthermore, our MSP analysis represented the hypermethylation of CDH1, SFRP1, and SFRP5 in HBV-HCCs, demonstrating an epigenetic silencing event directed by HBx. The repression of SFRP1 and CDH1 leads to the subcellular localization of β-catenin that results in more cell proliferation.
In this study, there was a lower rate of GSK3β mRNA level among HBV-HCC tumors, which was associated with some mutations in the protein-binding domain of HBx (I88F/M and L116V). This finding is in line with a previous study, showing that the mutants of HBx are related to the suppressed status of GSK3β (51). Although the mechanism of this downregulation is not yet completely understood, the activation of Src kinases, ERK kinase, or interaction with APC are suggested (10). However, our findings can be used for further investigation to obtain mechanistic evidence in different cell lines and/or mice models, which is the most significant limitation of the present work. The lake of protein expression assays is another limitation of this study that is significant for confirmation of gene expression findings, which is under investigation in a parallel project.
In conclusion, HBV has significant roles in the activation of the Wnt pathway, probably via HBx and its mutant forms that interact with host proteins, leading to the activation of the Wnt pathway. Among the studied genes, it seems that URG7 and GSK3β are the most prominent molecular targets of HBx in the Wnt pathway that can be elegant choices for future investigations. Regarding the transactivating and oncogenic potential of HBx that has significant roles in HBV replication and regulation of the Wnt pathway, it’s targeting as a therapeutic approach might have a promising perspective.