1. Background
Hepatitis C virus (HCV) is a leading cause of both acute and chronic hepatitis (1). According to a WHO recent report, the worldwide estimation of chronic hepatitis C virus-infected population was 71 million, which is around 1 % of the global population with the risk of developing cirrhosis and hepatocellular carcinoma (2). In Egypt, from 50 million people participated in the nationwide program to eliminate HCV, the seropositivity was 4.6% which was much higher than the global estimates (3).
In the patient’s serum, circulating HCV particles were embedded on the surfaces of host low density and very low density lipoproteins (LDL and VLDL) (4). These viral RNA-host lipoprotein complexes are called lipo-viro- particles (LVPs) (5).
The structure of APO E in purified LVPs has been shown with immunogold electron microscopy (6). It was confirmed to play an important role in the generation of infectious HCV particles (7). Severe reduction in HCV particle assembly and release and decreased HCV infectivity was reported following APO E knockdown proving an additional role for APO E in regulating virus assembly /release and in virus entry (8).
Apo E gene has three relatively common allelic variants, ApoE-ε2, ApoE-ε3, and ApoE-ε4, defined by two SNPs, rs429358 and rs7412. The most common variant overall is the “standard” ApoE-ε3 (9).
The impact of Apolipoprotein E isoforms on hepatitis C virus disease progression and response to interferon therapy had been addressed (10), but it has not yet been shown in patients receiving direct-acting antivirals.
2. Objectives
The present study was conducted to determine the impact of apolipoprotein E gene polymorphism on HCV disease progression and response to direct-acting antivirals in the Egyptian population.
3. Methods
We conducted a retrospective observational cohort study in the period from April 2017 to September 2017. Ninety-six patients with chronic hepatitis C were enrolled from Zagazig Viral Hepatitis Treatment Center, Sharkia governorate who completed antiviral therapy with SOF/DCV or SOF/DCV/RBV for 12 weeks. Twenty patients were selected from non-responders to DAAs treatment and 76 subjects were selected from responders. The exclusion criteria were patients with combined infections (HCV/HBV or HCV/HIV), hepatocellular carcinoma or other malignancies, previous treatment with interferon, and those patients using immunosuppressive therapy. The study was approved by the Institutional Review Board (IRB). The subjects were asked to sign the written consent; Full history taking, clinical examination, and initial data collection were performed for each subject. To assess the severity of the liver disease, the Child-Pugh score, MELD score, and Fib-4 were calculated.
3.1. Apo E Genotypes Determination
For genotyping of Apo E in patients, Genomic DNA was extracted from the whole blood sample using Gene JET Whole Blood DNA Purification Kit (Thermo Scientific). The Apo E gene was amplified via a nested polymerase chain reaction (PCR) using the primers and thermal profile in Table 1 to assess polymorphism.
| Primer Designation | Sequences (5 → 3) | Fragment Positiona | Product Size, bp | Denaturation | Annealing | Extension |
|---|---|---|---|---|---|---|
| Outer F | 5’TGAAGGAGTTGAAGGCCTAC3’ | 336 - 317 | 385 | 95°C for 30 sec | 53°C for 30 sec | 72°C for 1 min |
| Outer R | 5’CACGCGGCCCTGTTCCACCA3’ | 684 - 664 | ||||
| Inner F | 5’TCCAAGGAGCTGCAGGCGGCGCA3’ | 394 - 416 | 218 | 95°C for 30 sec | 55°C for 30 sec | 72°C for 30 sec |
| Inner R | 5’GCCCCGGCCTGGTACACTGCCA3’ | 611 - 589 |
aGenBank accession number NM_000041.1.
PCR was performed using Dream Taq Green PCR Master Mix (Thermo Scientific) on Veriti 96 Well Thermal Cycler (Applied Bio Systems). For the PCR reaction, an initial denaturation step was performed at 95°C for 3 minutes, followed by 35 cycles of denaturation, annealing, and extension based on the conditions in Table 1. A final extension step at 72°C for 5 minutes was performed. The different alleles of Apo E genotypes were determined after the digestion of each product using the AflIII (Thermo Fisher Scientific) and HaeII (BfoI) (New England Biological) restriction enzymes separately and running each on 3% agarose gel stained with a low concentration of ethidium bromide (11) (Table 2) (12).
| Apo E Genotype | AflIII Cut Bands | HaeII (BfoI) Cut Bands |
|---|---|---|
| E3/E3 homozygous alleles | 168 + 50 | 195 + 23 |
| E2/E2 homozygous alleles | 168 + 50 | 218 (No cut) |
| E4/E4 homozygous alleles | 218 (No cut) | 195 + 23 |
| E3/E4 heterozygous alleles | 218 + 168 + 50 | 195 + 23 |
| E2/E3 heterozygous alleles | 168 + 50 | 218 + 195 + 23 |
4. Results
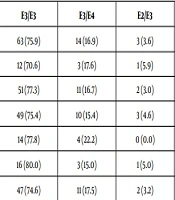
The study included 96 patients, 13 of them were excluded from the study due to incomplete results.. Apo E genotype E3/E3 was found to be the most common genotype in patients with chronic hepatitis C (CHC) (75.9% of cases) followed by genotype E3/E4 (16.9%). Other types such as E2/E3, E2/E2, E4/E4 were observed in a few cases (Table 3).
No statistically significant association between cirrhotic status, treatment response, viremia level of the patients and Apo E genotypes (Table 3).
| APO E Genotype | E3/E3 | E3/E4 | E2/E3 | E4/E4 | E2/E2 | χ2 | P |
|---|---|---|---|---|---|---|---|
| Total (n = 83) | 63 (75.9) | 14 (16.9) | 3 (3.6) | 2 (2.4) | 1 (1.2) | 4.8 | 0.3 |
| Cirrhotic (n = 17) | 12 (70.6) | 3 (17.6) | 1 (5.9) | 0 (0.0) | 1 (5.9) | ||
| Non-cirrhotic (n = 66) | 51 (77.3) | 11 (16.7) | 2 (3.0) | 2 (3.0) | 0 (0.0) | ||
| SVR (n = 65) | 49 (75.4) | 10 (15.4) | 3 (4.6) | 2 (3.1) | 1 (1.5) | 2.1 | 0.7 |
| Non-SVR (n = 18) | 14 (77.8) | 4 (22.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | ||
| High-viremia (n = 20) | 16 (80.0) | 3 (15.0) | 1 (5.0) | 0 (0.0) | 0 (0.0) | 1.2 | 0.8 |
| Low-viremia (n = 63) | 47 (74.6) | 11 (17.5) | 2 (3.2) | 2 (3.2) | 1 (1.6) |
aValues are expressed as No. (%).
5. Discussion
Chronic HCV infection remains a major global health concern, particularly in Egypt (13). In 2016, the WHO implemented a viral hepatitis elimination program. Many countries attempted to adopt this program in their health strategies agenda (14, 15). In this study, we evaluated the impact of apolipoprotein E gene polymorphism on HCV disease progression and response to DAA in Egypt.
In line with several epidemiological studies (16, 17), the Apo E3 allele was by far the most common allele encountered in our study, with a frequency of 86.1 %.
In our results, two patients with E4/E4 genotype were non cirrhotics had a sustained virologic response and were of low viremia. Although this may reflect a protective effect for the Apo E4 allele, the limited number yields a non-significant association (P = 0.08).
There was no statistically significant association between Apo E genotype and severity of liver disease (P = 0.3). Fabris et al. (12) reported that the carriage of the Apo E3/E3 genotype was associated with faster average fibrosis progression and that carriage of the Apo E4 and of the Apo E2 alleles were associated, in chronic hepatitis C, with less severe liver disease. However, this difference may be due to different study populations. Also, the limited number of E2 and E4 genotypes in this study could be a reason for the insignificant association.
In line with the results obtained by Wozinak et al. (18), no significant association was found between Apo E genotypes with the response to DAAs treatment. Our results and results by Wozinak et al. (18) were different from findings of Price et al. (19) who revealed that Apo E4 was associated with a low risk of viral persistence. Another study was conducted from Egypt, reported that the Apo E3 allele could be a risk marker for resistance to combined therapy (20).
It is necessary to identify the reasons for the diversity of HCV infection results. The present study faced a few limitations. One of the most obvious limitations was the small number of patients included in the study, so further studies are required on a larger number of patients to confirm the results of this study.
5.1. Conclusions
Although no statistically significant association was found between any of apolipoprotein E genotypes and progression of HCV disease or response to treatment with direct-acting antivirals, the limited number of E4 and E2 genotypes was a major limitation. These findings need to be confirmed by further prospective studies using larger number of patients.