1. Context
Coinfection of Hepatitis D virus (HDV) with Hepatitis B virus (HBV) increases the risk of progression to end-stage liver diseases such as cirrhosis and hepatocellular carcinoma (HCC) (1, 2). The prevalence of HDV infection in individuals with HBsAg varies by their clinical presentations of the disease (more prevalent in cirrhotic than in non-cirrhotic patients) and geographical location (high prevalence in South America, Eastern Europe, and Sub-Saharan Africa and low prevalence in Western Europe, North America, and Southeast Asia) (3, 4). Pegylated-interferon with 20% - 30% treatment success is the currently available standard of care for the management of HDV infection (5). So far, there is no effective treatment against hepatitis D infection. However, the hope for the development of an effective treatment for HDV infection in the near future has been strengthened in recent years (6).
Hepatitis D virus with a 1.7 kb circular RNA genome displays high genetic variability and traditionally is classified into three genotypes (HDV I, II, and III) (7). Hepatitis D virus genotype I is globally distributed, HDV genotype II is mainly detected in East and Southeast Asia, and HDV genotype III is exclusively observed in South America (8). However, a comprehensive phylogenetic analysis of HDV full-length sequences including African strains showed eight distinct clades (HDV-1 to -8), representing eight genotypes of HDV (7, 9). In addition to the different geographical distribution, HDV genotypes differ in prognosis, clinical presentation, progression, and response to therapy (10-12).
Despite the effective improvement of therapeutic and preventive interventions that have led to decreasing the burden of hepatitis B-related liver diseases, HBV/HDV management remained troublesome (13). In line with the goals of global elimination of viral hepatitis and parallel with the ongoing focus on hepatitis B and C viruses, HDV should not be neglected (14, 15). Regarding the different responses of HDV to antiviral therapy and coinfection with HBV, deep insight into the molecular epidemiology and global distribution of HDV genotypes fulfills the goal of the elimination program.
2. Objectives
This systematic review was conducted to present a clear picture of HDV genotypes dispersal at the global and regional levels.
3. Methods
3.1. Search Protocols, Data Resources, and Registration
Through the development of a specific search strategy in three major electronic databases including PubMed, Scopus, and Science Citation Index Expanded (Web of Science), we covered the main parts of our topic including “hepatitis D virus” and “HDV genotypes”. All protocols were peer-reviewed transversely and parallelly by all authors. The complete search protocols for each database are addressed in supplementary file Appendix 1. Initially, the search was performed on September 16, 2019. However, it was updated on February 2, 2020. Finally, references of all the included studies were screened for finding relevant titles. The protocol of the current systematic review has been registered in the International Prospective Register of Systematic Reviews (PROSPERO) under record identification CRD42018097296, available at: https://www.crd.york.ac.uk/PROSPERO/display_record.php?RecordID=97296.
3.2. Selection Criteria
We included all original articles whether clinical or molecular epidemiologic studies relevant to HDV and its genotypes. The inclusion criteria were studies on HDV genotyping using the molecular method on blood serum/plasma specimens, the minimum number of 10 cases (sample size) from each geographical region (country), and at least an English abstract. Review articles, letters, editorials, case reports, conference abstracts, and proceedings were excluded from further processing.
3.3. Quality Assessment, Study Selection, and Data Extraction
Based on the PRISMA guidelines for reporting systematic reviews (16), the quality of the included studies was assessed over the sampling method, coverage of the general population, and study level (regional, national, provincial, etc.). Using EndNote version 9, any duplicated item was removed. According to the eligibility criteria, two authors (MSR-Z and HS) screened all the studies independently at three levels of title, abstract, and full-text, in sequence. In the case of disagreement for including/excluding a study, it was resolved by mutual discussion between them or with other authors. The following data were extracted from each included study: First author’s name, publication year, HDV genotyping method, HDV genotype classification, sample size, the geographical location of sampling, and prevalence of each HDV genotype. The old HDV genotype classification (HDV I, II, and III) and African HDV genotypes (HDV-5, -6, -7, and -8) were used for showing the HDV genetic dispersion to have the chance to include all studies regardless of their HDV genotype classification. The results of the new HDV genotype classification (HDV-1 to -8) were converted to the old one (HDV I, II, and III) using the proposal for HDV genus classification (7).
3.4. Analysis
The distribution of HDV genotypes was reported in a table concerning the geographical location of sampling. When there was a possibility for pooling data, the distribution of HDV genotypes for each country was pooled and translated to its global and regional distribution.
4. Results
4.1. Study Screening
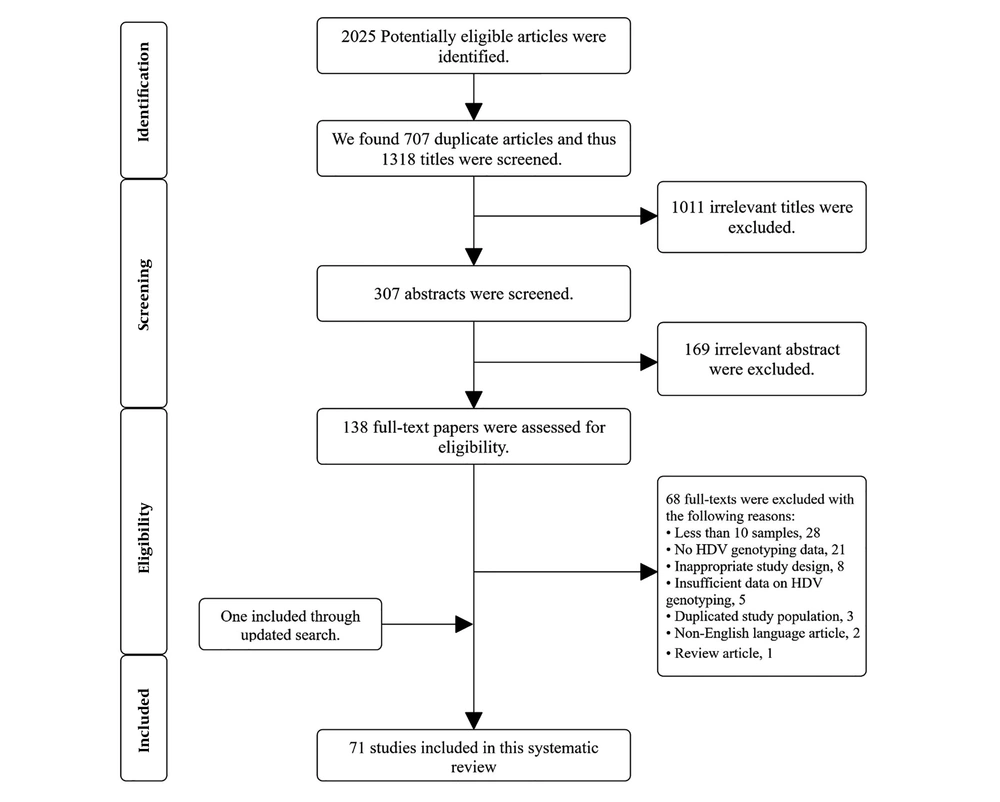
The main search on September 16, 2019, resulted in the recruitment of 2,025 published articles. After removing duplicates and screening of titles and abstracts, 138 full-texts were selected for inclusion in the current systematic review. The investigation was updated on February 2, 2020, and reached us to 71 studies on the evaluation of HDV genotypes for the final qualitative analysis. The complete process of screening and selection of articles is shown in Figure 1.
4.2. Characteristics of Included Studies
Among 71 studies, 43 (60.6%) used the new HDV genotype classification while 28 (39.4%) used the old HDV genotype classification. Among all, 52 (73.2%) applied sequencing as the HDV genotyping method, 10 (14.1%) used restriction fragment length polymorphism (RFLP), seven (9.9%) used sequencing and RFLP, and two (2.8%) used reverse hybridization. Data extraction of the 71 studies identified 77 records (based on the geographical location of sampling) in 33 countries. The sampling locations were Brazil (17-25) with nine records, Iran (26-33) with eight records, Turkey (34-39) and Taiwan (12, 40-44) each with six records, Italy (33, 45-47) and Pakistan (48-51) each with four records, Gabon (52-54), Mongolia (55-57), Nigeria (58-60), and Vietnam (61-63) each with three records, Cameroon (64, 65), Germany (33, 66), Mauritania (67, 68), Romania (47, 69), and Spain (33, 70) each with two records, and Albania (71), Australia (72), Central African Republic (73), China (74), Congo (75), Ethiopia (76), France (77), Greece (71), Israel (78), Japan (79), Kiribati (80), Peru (81), Russia (82), Tajikistan (83), Tunisia (84), UK (11), USA (71), and Venezuela (85) each with one record. The characteristics of the included studies are summarized in Table 1.
| Author and Reference | Publication Year | HDV Genotyping Method | HDV Genotype Classification | Sampling Location, No. | HDV Genotype, No (%) | Notes | |||
|---|---|---|---|---|---|---|---|---|---|
| HDV I (HDV-1) | HDV II (HDV-2 or HDV-4) | HDV III (HDV-3) | Othera | ||||||
| Wu et al. (40) | 1995 | RFLP | Old | Taiwan, 88 | 24 (27.3) | 64 (72.3) | 0 (0) | 0 (0) | |
| Casey et al. (81) | 1996 | RFLP | Old | Peru, 37 | 0 (0) | 0 (0) | 37 (100) | 0 (0) | |
| Niro et al. (45) | 1997 | Sequencing | Old | Italy, 46 | 46 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Shakil et al. (71) | 1997 | Sequencing | Old | Greece, 30 | 30 (100) | 0 (0) | 0 (0) | 0 (0) | |
| USA, 22 | 22 (100) | 0 (0) | 0 (0) | 0 (0) | |||||
| Albania, 10 | 10 (100) | 0 (0) | 0 (0) | 0 (0) | |||||
| Cotrina et al. (70) | 1998 | Sequencing | Old | Spain, 37 | 37 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Wu et al. (41) | 1998 | RFLP and sequencing | Old | Taiwan, 58 | 5 (8.6) | 53 (91.4) | 0 (0) | 0 (0) | Five samples presumed as HDV IIb |
| Ivaniushina et al. (82) | 2001 | RFLP and sequencing | Old | Russia, 29 | 14 (48.3) | 15 (51.7) | 0 (0) | 0 (0) | |
| Quintero et al. (85) | 2001 | Sequencing | Old | Venezuela, 12 | 7 (58.3) | 0 (0) | 5 (41.7) | 0 (0) | |
| Kao et al. (42) | 2002 | RFLP | Old | Taiwan, 31 | 11 (35.5) | 18 (58.1) | 0 (0) | 2 (6.4) | Two with mixed HDV I and HDV II infection |
| Watanabe et al. (79) | 2003 | Sequencing | Old | Japan, 33 | 2 (6.1) | 31 (93.9) | 0 (0) | 0 (0) | One HDV IIa and 30 HDV IIb |
| Bozdayi et al. (34) | 2004 | RFLP | Old | Turkey, 59 | 59 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Takahashi et al. (55) | 2004 | Sequencing | Old | Mongolia, 20 | 20 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Kaymakoglu et al. (35) | 2005 | RFLP | Old | Turkey, 19 | 19 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Tsatsralt-Od et al. (57) | 2005 | Sequencing | Old | Mongolia, 117 | 117 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Parana et al. (17) | 2006 | Reverse hybridization | Old | Brazil, 40 | 22 (55) | 0 (0) | 18 (45) | 0 (0) | |
| Su et al. (12) | 2006 | RFLP | New | Taiwan, 133 | 51 (39.1) | 82 (60.9) | 0 (0) | 0 (0) | 74 HDV IIa and eight HDV IIb |
| Tsatsralt-Od et al. (56) | 2006 | Sequencing | Old | Mongolia, 32 | 32 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Altuglu et al. (36) | 2007 | Sequencing | Old | Turkey, 26 | 26 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Moatter et al. (48) | 2007 | RFLP | Old | Pakistan, 23 | 23 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Gomes-Gouvea et al. (18) | 2008 | Sequencing | New | Brazil, 14 | 0 (0) | 0 (0) | 14 (100) | 0 (0) | |
| Khan et al. (83) | 2008 | Sequencing | Old | Tajikistan, 10 | 10 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Mohebbi et al. (26) | 2008 | Sequencing | New | Iran, 22 | 22 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Esmaeili et al. (27) | 2009 | Sequencing | New | Iran, 26 | 26 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Makuwa et al. (52) | 2009 | Sequencing | New | Gabon, 17 | 10 (58.8) | 0 (0) | 0 (0) | 7 (41.2) | Two HDV-7 and five HDV-8 |
| Mirshafiee et al. (28) | 2009 | RFLP | Old | Iran, 13 | 13 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Hofmann et al. (66) | 2010 | Sequencing | New | Germany, 42 | 42 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Chang et al. (43) | 2011 | Sequencing | New | Taiwan, 99 | 5 (5.1) | 94 (94.9) | 0 (0) | 0 (0) | 41 HDV-2, 51 HDV-4, and two HDV-2/HDV-4 mixed infection |
| Foupouapouognigni et al. (64) | 2011 | Sequencing | New | Cameroon, 25 | 22 (88) | 0 (0) | 0 (0) | 3 (12) | One HDV-5, one HCV-6, and one HDV-7 |
| Pollicino et al. (46) | 2011 | Sequencing | New | Italy, 21 | 21 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Le Gal et al. (37) | 2012 | Sequencing | New | Turkey, 34 | 34 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Mansour et al. (67) | 2012 | Sequencing | New | Mauritania, 28 | 25 (89.3) | 0 (0) | 0 (0) | 3 (10.7) | Three HDV-5 |
| Mansour et al. (68) | 2012 | Sequencing | New | Mauritania, 31 | 28 (90.3) | 0 (0) | 0 (0) | 3 (9.7) | Three HDV-5 |
| Perveen et al. (49) | 2012 | RFLP and sequencing | New | Pakistan, 22 | 22 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Ghamari et al. (29) | 2013 | Sequencing | New | Iran, 12 | 12 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Karaca et al. (38) | 2013 | RFLP | Old | Turkey, 32 | 32 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Popescu et al. (69) | 2013 | Sequencing | New | Romania, 40 | 40 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Sy et al. (61) | 2013 | Sequencing | New | Vietnam, 21 | 19 (90.5) | 2 (9.5) | 0 (0) | 0 (0) | Two HDV-2 |
| Andernach et al. (58) | 2014 | Sequencing | New | Nigeria, 15 | 8 (53.3) | 0 (0) | 0 (0) | 7 (46.7) | Five HDV-5, two HDV-6 |
| Braga et al. (19) | 2014 | Sequencing | New | Brazil, 28 | 0 (0) | 0 (0) | 28 (100) | 0 (0) | |
| Bulut et al. (39) | 2014 | RFLP and sequencing | Old | Turkey, 113 | 113 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Butt et al. (50) | 2014 | Sequencing | New | Pakistan, 21 | 21 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Crispim et al. (20) | 2014 | RFLP and sequencing | Old | Brazil, 38 | 0 (0) | 0 (0) | 38 (100) | 0 (0) | |
| Han et al. (80) | 2014 | Sequencing | New | Kiribati, 14 | 14 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Kay et al. (21) | 2014 | Reverse hybridization | Old | Brazil, 90 | 0 (0) | 0 (0) | 90 (100) | 0 (0) | |
| Servant-Delmas et al. (77) | 2014 | Sequencing | New | France, 14 | 12 (85.7) | 0 (0) | 0 (0) | 2 (14.3) | One HDV-6 and one HDV-7 |
| Botelho-Souza et al. (22) | 2015 | RFLP and sequencing | Old | Brazil, 52 | 4 (7.7) | 0 (0) | 48 (92.3) | 0 (0) | |
| Lin et al. (44) | 2015 | Sequencing | New | Taiwan, 153 | 13 (8.5) | 140 (91.5) | 0 (0) | 0 (0) | 54 HDV-2 and 86 HDV-4 |
| Sadeghian et al. (30) | 2015 | RFLP | Old | Iran, 12 | 10 (83.3) | 2 (16.7) | 0 (0) | 0 (0) | |
| Shirvani-Dastgerdi et al. (31) | 2015 | Sequencing | New | Iran, 34 | 34 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Yacoubi et al. (84) | 2015 | Sequencing | New | Tunisia, 11 | 11 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Borzacov et al. (23) | 2016 | Sequencing | New | Brazil, 22 | 0 (0) | 0 (0) | 22 (100) | 0 (0) | |
| Francois-Souquiere et al. (53) | 2016 | Sequencing | New | Gabon, 16 | 2 (12.5) | 0 (0) | 0 (0) | 14 (87.5) | 14 HDV-8 |
| Opaleye et al. (59) | 2016 | Sequencing | New | Nigeria, 14 | 14 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Chen et al. (74) | 2017 | Sequencing | Old | China, 48 | 0 (0) | 48 (100) | 0 (0) | 0 (0) | |
| Makiala-Mandanda, et al. (75) | 2017 | Sequencing | New | Congo, 12 | 12 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Nguyen et al. (62) | 2017 | Sequencing | New | Vietnam, 25 | 5 (20) | 20 (80) | 0 (0) | 0 (0) | |
| Aberra et al. (76) | 2018 | Sequencing | New | Ethiopia, 12 | 12 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Aftab et al. (51) | 2018 | Sequencing | New | Pakistan, 17 | 17 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Binh et al. (63) | 2018 | Sequencing | New | Vietnam, 57 | 52 (91.2) | 5 (8.8) | 0 (0) | 0 (0) | |
| Butler et al. (65) | 2018 | Sequencing | New | Cameroon, 211 | 138 (65.4) | 0 (0) | 0 (0) | 73 (34.6) | 11 HDV-6, 61 HDV-7, one HDV-8 |
| Jackson et al. (72) | 2018 | Sequencing | New | Australia, 169 | 136 (80.5) | 6 (3.5) | 0 (0) | 27 (16) | 27 HDV-5 |
| Komas et al. (73) | 2018 | Sequencing | New | Central African Republic, 18 | 18 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Meysami et al. (32) | 2018 | RFLP and sequencing | Old | Iran, 37 | 32 (86.5) | 3 (8.1) | 0 (0) | 2 (5.4) | Two indeterminate isolates; sequencing of one indeterminate and one HDV II isolate resulted in HDV-1 genotypes. |
| Ricco et al. (47) | 2018 | Sequencing | New | Italy, 69 | 69 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Romania, 53 | 53 (100) | 0 (0) | 0 (0) | 0 (0) | |||||
| Shirazi et al. (78) | 2018 | Sequencing | New | Israel, 55 | 55 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Da Silva et al. (24) | 2019 | Sequencing | New | Brazil, 54 | 4 (7.4) | 0 (0) | 48 (88.9) | 2 (3.7) | Two HDV-5 |
| Groc et al. (54) | 2019 | Sequencing | New | Gabon, 42 | 23 (54.8) | 0 (0) | 0 (0) | 19 (45.2) | Seven HDV-7 and 12 HDV-8 |
| Ifeorah et al. (60) | 2019 | Sequencing | New | Nigeria, 10 | 9 (90) | 0 (0) | 0 (0) | 1 (10) | One HDV-6 |
| Karimzadeh et al. (33) | 2019 | Sequencing | New | Germany, 72 | 72 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Italy, 18 | 18 (100) | 0 (0) | 0 (0) | 0 (0) | |||||
| Spain, 12 | 12 (100) | 0 (0) | 0 (0) | 0 (0) | |||||
| Iran, 14 | 14 (100) | 0 (0) | 0 (0) | 0 (0) | |||||
| Scarponi et al. (25) | 2019 | Sequencing | New | Brazil, 26 | 26 (100) | 0 (0) | 0 (0) | 0 (0) | |
| Spaan et al. (11) | 2020 | Sequencing | New | UK, 39 | 18 (46.2) | 0 (0) | 0 (0) | 21 (53.8) | 21 HDV-5 |
aHDV-5, HDV-6, HDV-7, HDV-8, mixed infection, and unknown genotypes.
4.3. Country-Level, Regional, and Global Distribution of HDV Genotypes
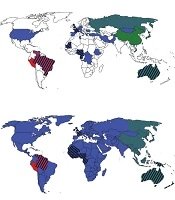
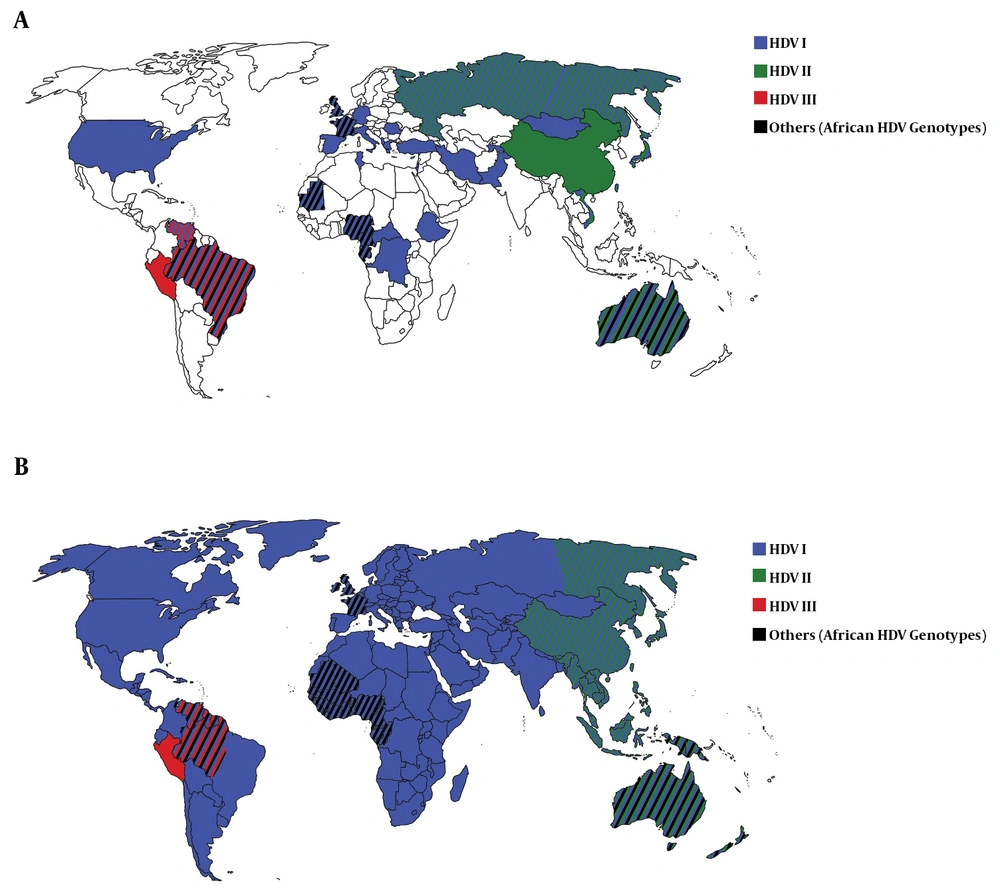
In Africa, the most common HDV genotype was HDV I. In the Central African Republic, Congo, Ethiopia, and Tunisia, all HDV isolates belonged to HDV I. In the West African countries including Cameroon, Gabon, Mauritania, and Nigeria, in addition to HDV I, African HDV genotypes (HDV-5, -6, -7, and -8) were observed (Table 2 and Figure 2A). In Africa and among African HDV genotypes, HDV-5 was isolated from patients in Cameroon, Mauritania, and Nigeria, HDV-6 from patients in Cameroon and Nigeria, and HDV-7 and -8 from patients in Cameroon and Gabon.
The most frequent HDV genotype in Asia was HDV II, followed by HDV I. In China, Japan, and Taiwan, the most common isolate was HDV II and in Mongolia, Pakistan, Tajikistan, and Vietnam, the most common isolate was HDV I. In Asia, no HDV III and African genotypes were detected (Table 2 and Figure 2A).
Among European countries, the most detected HDV genotype was HDV I so that in Albania, Germany, Greece, Italy, Romania, and Spain, only HDV I was identified from patients with HDV infection. In France and the UK, in addition to HDV I, African HDV genotypes were observed, as well. In Russia, both HDV I and HDV II were isolated (Table 2 and Figure 2A).
In the Middle Eastern countries, the most common HDV genotype was HDV I. Israel and Turkey observed only HDV I among patients with HDV infection while few HDV II isolates (genotyped by RFLP) were reported in Iranian patients (Table 2 and Figure 2A).
Only one study over HDV genotypes (with 22 isolates) was found from North America. All these isolates belonged to HDV I. Two studies from Oceania identified only HDV I in Kiribati while HDV I and II and African HDV genotypes (HDV-5) were observed in Australia (Table 2 and Figure 2A).
In South America, the most common HDV genotype was HDV III, followed by HDV I. In Peru, only HDV III was isolated from patients with HDV infection while in Venezuela, both HDV I and III were observed. In the north of Brazil, the most common HDV genotype was HDV III. In addition, a proportion of patients were infected with HDV I and a few numbers were infected with African HDV genotypes (HDV-5). In the south of Brazil, all HDV isolates belonged to HDV I (Table 2 and Figure 2A).
Worldwide, HDV I had global dispersion, HDV II was observed mainly in East Asia and Oceania, and HDV III was reported exclusively in the north of South America. African HDV genotypes were observed mainly in the west of Africa although these genotypes were isolated in Europe, South America, and Australia, as well (Table 2 and Figure 2A).
| Region, Country | Records (N) | Isolates (N) | HDV Genotypesa | |||
|---|---|---|---|---|---|---|
| HDV I (HDV-1) | HDV II (HDV-2 or HDV-4) | HDV III (HDV-3) | Others (HDV-5, -6, -7, or -8) | |||
| Africa | ||||||
| Cameroon | 2 | 236 | High | Very low | Very low | Intermediate |
| The Central African Republic | 1 | 18 | Very high | Very low | Very low | Very low |
| Congo | 1 | 12 | Very high | Very low | Very low | Very low |
| Ethiopia | 1 | 12 | Very high | Very low | Very low | Very low |
| Gabon | 3 | 75 | Intermediate | Very low | Very low | High |
| Mauritania | 2 | 59 | High | Very low | Very low | Low |
| Nigeria | 3 | 39 | High | Very low | Very low | Intermediate |
| Tunisia | 1 | 11 | Very high | Very low | Very low | Very low |
| Total | 14 | 462 | High | Very low | Very low | Intermediate |
| Asia | ||||||
| China | 1 | 48 | Very low | Very high | Very low | Very low |
| Japan | 1 | 33 | Low | High | Very low | Very low |
| Mongolia | 3 | 169 | Very high | Very low | Very low | Very low |
| Pakistan | 4 | 83 | Very high | Very low | Very low | Very low |
| Taiwan | 6 | 560 | Intermediate | High | Very low | Very low |
| Tajikistan | 1 | 10 | Very high | Very low | Very low | Very low |
| Vietnam | 3 | 103 | High | Intermediate | Very low | Very low |
| Total | 19 | 1006 | Intermediate | High | Very low | Very low |
| Europe | ||||||
| Albania | 1 | 10 | Very high | Very low | Very low | Very low |
| France | 1 | 14 | High | Very low | Very low | Low |
| Germany | 2 | 114 | Very high | Very low | Very low | Very low |
| Greece | 1 | 30 | Very high | Very low | Very low | Very low |
| Italy | 4 | 154 | Very high | Very low | Very low | Very low |
| Romania | 2 | 93 | Very high | Very low | Very low | Very low |
| Russia | 1 | 29 | Intermediate | High | Very low | Very low |
| Spain | 2 | 49 | Very high | Very low | Very low | Very low |
| UK | 1 | 39 | Intermediate | Very low | Very low | High |
| Total | 15 | 532 | High | Low | Very low | Low |
| Middle East | ||||||
| Iran | 8 | 168 | High | Low | Very low | Very low |
| Israel | 1 | 55 | Very high | Very low | Very low | Very low |
| Turkey | 6 | 283 | Very high | Very low | Very low | Very low |
| Total | 15 | 506 | Very high | Low | Very low | Very low |
| North America | ||||||
| US | 1 | 22 | Very high | Very low | Very low | Very low |
| Total | 1 | 22 | Very high | Very low | Very low | Very low |
| Oceania | ||||||
| Australia | 1 | 169 | High | Low | Very low | Intermediate |
| Kiribati | 1 | 14 | Very high | Very low | Very low | Very low |
| Total | 2 | 183 | High | Low | Very low | Low |
| South America | ||||||
| Brazil | 9 | 364 | Intermediate | Very low | High | Very low |
| Peru | 1 | 37 | Very low | Very low | Very high | Very low |
| Venezuela | 1 | 12 | High | Very low | Intermediate | Very low |
| Total | 11 | 413 | Intermediate | Very low | High | Very low |
| World (Total) | 77 | 3124 | High | Intermediate | Low | Low |
aPrevalence of HDV genotypes: < 1%, very low; 1 - 14.9%, low; 15 - 49.9%, intermediate; 50 - 98.9%, high; ≥ 99%, very high.
5. Conclusions
This systematic review highlights the global distribution of HDV genotypes, showing that HDV I has a global distribution and HDV II is mainly observed in East Asia. Accordingly, HDV III exclusively is circulating in South America and African HDV genotypes are majorly isolated in West Africa. Still, understanding the molecular epidemiology of infectious diseases such as HBV/HDV coinfection is crucial for vaccine and antiviral agent development and optimal management of the disease (86, 87).
In Africa, HDV I and African HDV genotypes were isolated from patients with HDV infection (52-54, 58-60, 64, 65, 67, 68, 73, 75, 76, 84). Based on the collected data, it seems that the African HDV genotypes originated from western countries of Africa such as Gabon, Cameroon, and Nigeria where the prevalence of HDV infection is high (53, 64). In Asia, both HDV I and HDV II were the predominant genotypes in patients with HDV infection (12, 40-44, 48-51, 55-57, 61-63, 74, 79, 83). Hepatitis D virus II was mainly observed in China (74), Taiwan (12, 40-44), and Japan (79) while in other Asian countries such as Mongolia (55-57), Pakistan (48-51), Tajikistan (83), and Vietnam (61-63), HDV I was the main genotype isolated from patients. The latter finding confirms that HDV II originated from the Far East. In European countries, HDV I, II, and African genotypes were detected in patients with HDV infection (11, 33, 45-47, 66, 69-71, 77, 82). In detail, in Albania (71), Germany (33, 66), Greece (71), Italy (33, 45-47), Romania (47, 69), and Spain (33, 70), only HDV I was isolated. In the study by Ivaniushina et al. (82) in Russia, in addition to HDV I, HDV II was observed, which is justified with the fact that the study was conducted in Yakutia located in the east of Russia, a region near the Chinese borders. In a study by Servant-Delmas et al. (77) in France, among 14 HDV-infected patients, 12 were infected with HDV I and two with African HDV genotypes who originated from Sub-Saharan Africa where African HDV genotypes are prevalent. In another study by Spaan et al. (11) from the UK, 18 were infected with HDV I and 21 with African HDV genotypes. All those patients with African HDV genotype infections in this study were immigrants who originated from Africa. These two recent studies are important since they reported the introduction of new viral strains to Europe by immigration as previously discussed (88, 89). In the Middle East, mainly HDV I was isolated from patients with HDV infection (26-39, 78). Studies from Turkey (34-39) and Israel (78) exclusively detected HDV I. All Iranian studies (26-29, 31, 33), except two studies by Sadeghian et al. (30) and Meysami et al. (32), found HDV I as the only circulating HDV genotype in Iran. The study by Sadeghian et al. (30) using RFLP found 10 patients with HDV I and two with HDV II; however, these two cases with unexpected HDV genotype II were not confirmed by sequencing. In a study by Meysami et al. (32) using RFLP, 32 patients were genotyped as HDV I, three as HDV II, and two as indeterminate isolates. Sequencing of one indeterminate and one HDV II isolate resulted in HDV I; thus, the authors showed the aberrant result by RFLP in these two cases that may be due to unexpected polymorphisms in the restriction sites of SmaI on the HDV genome used for genotyping (32). In North America, the only available study by Shakil et al. (71) showed HDV I as the only circulating HDV genotype in the USA and North America. In Oceania, two studies from Australia (72) and Kiribati (80) found HDV I as the main HDV genotype isolated from patients with HDV infection; however, in Australia, HDV II and African HDV genotypes were found, as well. In this study (72), the two patients with HDV II originated from Vietnam and all 27 patients with African HDV genotypes originated from Western to Central African countries such as Sierra Leone, Liberia, and Nigeria. In South America, HDV III was the most prevalent isolate found in patients with HDV infection (17-25, 81, 85); however, HDV I and African HDV genotypes were isolated, as well. In Peruvian Amazon, HDV III was the only HDV genotype detected in HDV-infected patients (81). In Venezuela, both HDV I and HDV III were isolated from patients with HDV infection (85). In Brazilian Amazon, the most detected HDV genotype was HDV III (17-24) while in Minas Gerais state, south of Brazil, and outside of Amazon basin, the only isolated HDV genotype was HDV I (25). These results confirm Amazon as the origin of HDV III.
The main limitation of the current systematic review was the unavailability of eligible studies for many countries. Moreover, the two different HDV genotype classifications compelled us to transform the data of studies from new HDV genotype classification to the old one. Another limitation was the validity of RFLP for HDV genotyping, as few studies showed the aberrant results obtained using RFLP.
The current systematic review collected the available country-level evidence on the distribution of HDV genotypes and translated it to the regional and global distribution of HDV genotypes. With a global distribution of HDV I, we concluded that HDV II is mainly observed in East Asia and HDV III in South America while African genotypes are majorly detected in West Africa (Figure 2B). We also confirmed that immigration has a great role in changing the pattern of HDV genetic pool worldwide.