1. Background
Hepatocellular carcinoma (HCC) is the fifth most common cancer. Hepatocarcinogenesis is a process that involves many genes (1). It is believed that the most altering events in the growth of the disease are caused by tumor suppressor genes, oncogenes, and reactivation of developmental pathways and their related receptors. Data show that the development and progression of HCC is a multistage process (2). Most of the time, an underlying disease leads to HCC, which can be cirrhosis, chronic hepatitis B virus (HBV), or hepatitis C virus (HCV) infections. Viral hepatitis infections play an important role in end-stage liver disease, and the need for transplantation increases the risk of HCC, especially after chronic HBV and HCV pathogenesis (3, 4). Hepatocellular carcinoma can cause hepatocyte turnover, inflammation, and oxidative DNA damage (5). Although novel therapeutic strategies have been developed, the treatment of advanced HCC has a poor prognosis (6).
MicroRNAs (miRNAs) are short and non-coding RNAs that have 20 - 24 nucleotides and play an important role in biological processes through the post-transcriptional regulation of protein-coding genes (7, 8). Discovering miRNAs that have roles in hepatocarcinogenesis represents an important area of investigation (9, 10).
Single nucleotide polymorphisms (SNPs) are one of the most common types of genetic variations in the human genome. It has been well established that SNPs in the protein-coding genes can affect protein functions that, in turn, influences the individual susceptibility to cancer, and SNPs in miRNA might also contribute to cancer development (11). Open reading frames of miRNAs might have genetic polymorphisms and mutations that have roles in the outcome of liver transplantation. Therefore, the identification of genetic variations can help improve the clinical management of HCC (7, 12).
Evidence suggests that miRNAs are frequently deregulated in HCC, and some of them are related to clinicopathological aspects of HCC and post-liver transplant outcomes. Hepatocellular carcinoma was introduced as the deregulator of many critical genes’ expression in important cellular processes, such as cell cycle control, growth, migration, and apoptosis. On the other hand, miRNAs also interfere with many physiologic and pathologic processes. Besides, miRNA dysfunction is common in human disorders, including cancers. In cancers, miRNAs have different roles as tumor suppressors or oncogenes during their development (13). Identifying these genetic biomarkers can be helpful in the reduction of HCC liver transplant rejection through early diagnosis (14-16). It is worth noting that the role of the miRNA-146a polymorphism has been detected in the increased susceptibility to HCC (5, 17). Also, it is documented that the miRNA-196a-2 polymorphism is associated with HCC risk and it is higher in HBV-positive HCC patients (18-22). The genetic polymorphism of miRNA-149 is also associated with HBV-related HCC patients (18, 23), and the miRNA-499 polymorphism is important in patients with HCC (23, 24).
An SNP in the miRNA sequence might be a reason for alterations in miRNA expression and/or maturation, which is associated with cancer development and progression. MiRNA-196a-2 was mapped on chromosome 12q13.13, and an SNP was identified in its sequence (C > T; rs11614913). Several reports have identified miRNA-196a-2 (rs11614913) as a possible biomarker associated with multiple malignant tumors. Molecular epidemiologic studies have suggested that the polymorphism in miRNA-196a-2 (rs11614913) is associated with the increased risk of non-small cell lung cancer (NSCLC), breast cancer, HCC, gastric cancer, and head and neck cancer (25-27). The miRNA-146 was classified as miRNA-146a and miRNA-146b that overexpressed in HCC (26). A SNP called miRNA-146a (G > C, ch5q33; rs2910164) was found in miRNA-146a. The miRNA-146a (rs2910164) polymorphism is located in the stem region opposite to the mature miRNA-146a sequence. The optimal free energy was lower for the C allele than for the G allele, suggesting a less stable secondary structure for the C allele. The pre-microRNAs genetic variation of miRNA-146 changes the secondary structure and alters the expression of mature miRNAs (28).
MiRNA-499 (A > G, ch20q11.2; rs3746444) is another polymorphism, which is located within the 20th intron of the beta-myosin heavy chain gene with an essential role in the progression of cell proliferation (24). The miRNA-149 (C > T, ch2q37.3; rs2292832) was reported to be associated with a variety of malignancies, particularly digestive and prostatic cancers (18, 28, 29).
2. Objectives
Therefore, in continuation of our earlier report (30), in this study, the prevalence of miRNA-146a (rs2910164), miRNA-499 (rs3746444), miRNA-149 (rs2292832), and miRNA-196a-2 (rs11614913) gene polymorphisms were evaluated in liver recipients with HCC with or without experiencing graft rejection.
3. Methods
3.1. Study Population
A total of 60 tissue samples were selected from the sample bank of the pathobiology lab, collected from HCC patients who underwent liver transplant surgery at the Transplant Unit, Namazi Hospital, Shiraz, Iran, in 2013 - 2015. The control group consisted of 120 individuals randomly selected from the blood transfusion organization center, but those with a history of cancer or other diseases were excluded. Based on underlying diseases leading to HCC, the patients were subdivided into viral (HBV or HCV-related), and non-viral HCC. The underlying disease in most of the patients with non-viral HCC was cryptogenic cirrhosis (Table 1). The Local Ethics Committee of Shiraz University of Medical Sciences approved this study. Written informed consent was obtained from participants or their parents/guardians (for patients below 16-years-old). All patients and controls had Iranian nationality.
| Characteristics | HCC Patients | Rejected Group | Non-Rejected Group |
|---|---|---|---|
| Mean age | 45.23 ± 16.31 | 45.29 ± 16.73 | 45.22 ± 16.37 |
| Male | 47 (78.3) | 12 | 35 |
| Female | 23 (21.7) | 2 | 11 |
| Underlying disease, % | |||
| HBV-related HCC | 16 (26.7) | 4 (6.7) | 12 (20) |
| HCV-related HCC | 3 (5) | - | 3 (5) |
| Non-viral HCC | 41 (68.3) | 10 (16.7) | 31 (51.6) |
| Total | 60 (100) | 14 (23.4) | 46 (76.6) |
Abbreviations: HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HCV, hepatitis C virus.
aValues are expressed as No. (%) or mean ± SD.
3.2. DNA Extraction
The DNA was extracted from tissue samples using a DNA extraction kit or DNPTM kit (Cinna Gen, Iran) according to the manufacturer’s instruction. The DNA of control blood samples was extracted using the phenol-chloroform (Cinna Gen, Iran) extraction method according to the manufacturer’s instruction.
3.3. Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP) Method
SNPs of the studied genes: miRNA-146a (rs2910164), miRNA-499 (rs3746444), miRNA-149 (rs2292832), and miRNA-196a-2 (rs11614913) were analyzed by the PCR-RFLP method using a thermal cycler (Eppendorf, Germany). The primers, fragment sizes, and restriction enzymes used to evaluate the miRNA genetic polymorphisms are shown in Table 2.
| Genes (variants) | Primer Sequence (5’ → 3’) | Digestion Pattern/Fragment Sizes, bp | Restriction Enzymes |
|---|---|---|---|
| MiRNA-146aG/C | F: 5’-CATGGGTTGTGTCAGTGTCAGAGCT-3’ | GG = 147; CG = 147,122,25; CC = 122,25 | SacI (5’-G A G C T↓C-3’) |
| R:5’-TGCCTTCTGTCTCCAGTCTTCCAA-3’ | |||
| MiRNA-149C/T | F:5’-CTGGCTCCGTGTCTTCACTC-3’ | TT = 224,66; CT = 224,153,71,66; CC = 153,71 | AluI (5’- A G↓C T-3’) |
| R:5’-TGAGGCCCGAAACACCCGTA-3 | |||
| MiRNA-499A/G | F:5’-CAAAGTCTTCACTTCCCTGCCA-3’ | GG = 146; AG = 146,120,26; AA = 120,26 | BclI (5’- T↓G A T C A-3’) |
| R:5’-GATGTTTAACTCCTCTCCACGTGATC-3’ | |||
| MiRNA-196a2C/T | F:5’-CCCCTTCCCTTCTCCTCCAGATA-3-3’ | TT = 149; CT = 149,125,24; CC = 125,24 | MspI (5’-C↓C G G-3’) |
| R:5’-CGAAAACCGACTGATGTAACTCCG |
The PCR conditions for miRNA-146a (rs2910164) and miRNA-149 (rs2292832) were as follows: 95°C for 5 min, 40 cycles (95°C for 1 min, 61°C for 1 min, and 72°C for 1 min), and 72°C for 5 min. For miRNA-499 (rs3746444) and miRNA-196a-2 (rs11614913), the conditions were the same except that the annealing temperature was 59°C. The PCR mixture for amplification included PCR buffer (10×)/2.5 µL, MgCl2 (25 mM)/1.5 µL, dNTPs (10 mM)/0.5 µL, primers (10 pmol)/0.5 µL, Taq hot-start polymerase (5 units)/0.25 µL, and DNA/10 µL, which was the same for all samples.
3.4. Statistical Analysis
Direct gene counting was used to calculate the allele and genotype frequencies in patients and controls. Statistical analysis was carried out using SPSS Ver.16 for Windows (SPSS Inc., Chicago, IL, USA) and Epi Info software (CDC, Atlanta, USA). The frequencies of the alleles/genotypes were compared in both case and control groups by the χ2 test or Fisher’s exact test when appropriate. The statistical significance was set at the P value of less than 0.05 and calculated by the two-tailed method. The Hardy-Weinberg equilibrium of the studied alleles was evaluated by Arlequin ver.3.1.1 software (http://cmpg.unibe.ch/software/arlequin3).
4. Results
4.1. Patient Profiles
Among 60 recipients, the male to female ratio was 12/2 (6) in the rejected group and 35/11 (3.18) in the non-rejected group. The age range of the patients was 10 - 73 years, with a mean of 45.23 ± 16.31 years. The demographic data of the studied patients are shown in Table 1. In the present study, 8% of the recipients received grafts from living donors while the remaining 92% received them from cadavers. The control group consisted of 120 individuals (58.3% males). The average age was 30 - 60 years, with a mean of 45.09 ± 9.03 years.
The allele and genotype frequencies of miRNA-146a (rs2910164), miRNA-499 (rs3746444), miRNA-149 (rs2292832), and miRNA-196a-2 (rs11614913) were determined in the rejected and non-rejected groups of liver transplant recipients. Except for the miRNA-196a-2C > T genotype, all other studied miRNA genotypes in both groups of patients agreed with the Hardy-Weinberg equilibrium. The most frequent age range of patients was 50 - 65 years.
4.2. Comparing miRNA-146a (rs2910164), miRNA-499 (rs3746444), miRNA-149 (rs2292832), and miRNA-196a-2 (rs11614913) Polymorphisms in Patients and Controls
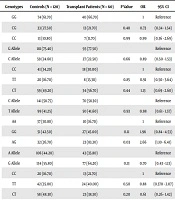
The AG genotype of the miRNA-499 (rs3746444) polymorphism was significantly lower in liver transplant patients than in controls (OR = 2.66, 95% CI: 1.10 - 6.41, P = 0.03). However, the genotypes and alleles of the other studied miRNA polymorphisms had no significant effect on the HCC outcomes of liver transplant recipients (Table 3).
| SNPs | Genotypes | Controls (N = 120) | Transplant Patients (N = 60) | P Value | OR | 95% CI |
|---|---|---|---|---|---|---|
| MiRNA-146G > C | GG | 74 (61.70) | 40 (66.70) | 1 | Reference | |
| CG | 33 (27.50) | 13 (21.70) | 0.40 | 0.73 | (0.34 - 1.54) | |
| CC | 13 (10.80) | 7 (11.70) | 0.99 | 0.99 | (0.36 - 2.69) | |
| G Allele | 181 (75.40) | 93 (77.50) | Reference | |||
| C Allele | 59 (24.60) | 27 (22.50) | 0.66 | 0.89 | (0.50 - 1.53) | |
| MiRNA-196a2C > T | CC | 41 (34.20) | 18 (30.00) | 1 | Reference | |
| TT | 20 (16.70) | 8 (13.30) | 0.85 | 0.91 | (0.50 - 3.64) | |
| CT | 59 (49.20) | 34 (56.70) | 0.44 | 1.13 | (0.69 - 2.60) | |
| C Allele | 141 (58.75) | 70 (58.30) | Reference | |||
| T Allele | 99 (41.25) | 50 (41.60) | 0.93 | 0.98 | (0.61 - 1.57) | |
| MiRNA-499A > G | AA | 37 (30.80) | 10 (16.70) | 1 | Reference | |
| GG | 51 (42.50) | 27 (45.00) | 0.11 | 1.96 | (0.84 - 4.53) | |
| AG | 32 (26.70) | 23 (83.30) | 0.03 | 2.66 | (1.10 - 6.41) | |
| A Allele | 106 (44.20) | 43 (35.80) | Reference | |||
| G Allele | 134 (55.80) | 77 (64.20) | 0.13 | 0.70 | (0.43 - 1.13) | |
| MiRNA-149C > T | CC | 20 (16.70) | 13 (21.70) | 1 | Reference | |
| TT | 42 (35.00) | 24 (40.00) | 0.50 | 0.88 | (0.370 - 2.07) | |
| CT | 58 (48.30) | 23 (38.30) | 0.20 | 0.61 | (0.26 - 1.42) | |
| C Allele | 98 (40.80) | 49 (40.80) | Reference | |||
| T Allele | 142 (59.20) | 71 (59.20) | 1.00 | 1.00 | (0.62 - 1.59) |
aValues are expressed as No. (%).
4.3. Inheritance of miRNA-146a (rs2910164), miRNA-499 (rs3746444), miRNA-149 (rs2292832), and miRNA-196a-2 (rs11614913) Genes in Transplant Recipients
The CC genotype and C allele of the miRNA-146a (rs2910164) polymorphism were associated with the increased risk of transplant rejection in HCC-disordered liver transplant patients (OR = 5.33, 95% CI: 0.98 - 28.77, P = 0.05; OR = 0.41, 95% CI: 0.15 - 1.15, P = 0.05, respectively). Furthermore, the C allele and CC genotype of miRNA-146a (rs2910164) were significantly more frequent in male patients who experienced acute rejection than in non-rejected patients (OR= 5.33, 95% CI: 0.78 - 39.66, P = 0.05; OR = 2.86, 95% CI: 0.94 - 8.74, P = 0.03, respectively). However, the alleles and genotypes of all studied miRNA polymorphisms had no significant effect on the HCC outcomes of liver transplant recipients (Table 4).
| SNPs | Genotypes | Rejected Group (N = 14) | Non-Rejected Group (N = 46) | P Value | OR | 95%CI |
|---|---|---|---|---|---|---|
| MiRNA-146G > C | GG | 8 (57.1) | 32 (69.6) | 1 | Reference | |
| CG | 2 (14.3) | 11 (23.9) | 0.71 | 0.72 | (0.13 - 3.95) | |
| CC | 4 (28.6) | 3 (6.5) | 0.05 | 5.33 | (0.98 - 28.77) | |
| G Allele | 18 (64.3) | 75 (81.5) | Reference | |||
| C Allele | 10 (35.7) | 17 (18.5) | 0.05 | 0.41 | (0.15 - 1.15) | |
| MiRNA-196a2C > T | CC | 6 (42.9) | 12 (26.1) | 1 | Reference | |
| TT | 1 (7.1) | 7 (15.2) | 0.28 | 0.28 | (0.02 - 2.88) | |
| CT | 7 (50) | 27 (58.7) | 0.31 | 0.51 | (0.14 - 1.87) | |
| C Allele | 19 (67.9) | 51 (55.4) | Reference | |||
| T Allele | 9 (32.1) | 41 (44.6) | 0.24 | 0.59 | (0.22 - 1.56) | |
| MiRNA-499A > G | AA | 2 (14.3) | 8 (17.4) | 1 | Reference | |
| GG | 7 (50) | 20 (43.5) | 0.71 | 1.40 | (0.23 - 8.24) | |
| AG | 5 (35.7) | 18 (39.1) | 0.91 | 1.11 | (0.17 - 6.99) | |
| A Allele | 9 (32.1) | 34 (37) | Reference | |||
| G Allele | 19 (67.9) | 58 (63) | 0.64 | 0.84 | (0.30 - 2.16) | |
| MiRNA-149C > T | CC | 3 (21.4) | 54 (58.7) | 1 | Reference | |
| TT | 6 (42.9) | 17 (37) | 0.69 | 0.90 | (0.23 - 5.4) | |
| CT | 5 (35.7) | 20 (43.5) | 0.60 | 0.93 | (0.18 - 4.70) | |
| C Allele | 11 (39.3) | 38 (41.3) | Reference | |||
| T Allele | 17 (60.7) | 9 (19.5) | 0.84 | 1.08 | (0.42 - 2.80) |
4.4. Comparing miRNA-146a (rs2910164), miRNA-499 (rs3746444), miRNA-149 (rs2292832), and miRNA-196a-2 (rs11614913) Polymorphisms in Viral Infected and Non-Viral Liver Transplant Patients
The C allele and CC genotype of the miRNA-146a (rs2910164) polymorphism were associated with the increased risk of transplant rejection in non-viral HCC liver transplant patients compared to non-rejected ones (OR = 6.21, 95% CI: 0.65 - 69.1, P = 0.04; OR = 3.71, 95% CI: 0.86 - 11.76, P = 0.04, respectively). However, the genotypes and alleles of the miRNA-499 (rs3746444), miRNA-149 (rs2292832), and miRNA-196a-2 (rs11614913) polymorphisms had no significant differences between non-viral-infected liver transplant recipients experiencing rejection and non-rejected ones. No statistically significant differences were found in the allele or genotype distributions of the studied miRNA polymorphisms, including miRNA-146a (rs2910164), miRNA-499 (rs3746444), miRNA-149 (rs2292832), and miRNA-196a-2 (rs11614913) between viral-infected liver transplant recipients experiencing rejection and viral infected non-rejected ones.
5. Discussion
The miRNA genes, located in cancer-associated genomic regions that might function either as tumor suppressors or oncogenes, can lead to malignant transformation (13, 17, 31). MiRNAs have roles in introducing and developing cancers and can be used to predict the prognosis or treatment response (32, 33). In HCC, an individual’s susceptibility to transplant rejection might be determined by genetic polymorphisms of the genes involved in multistage hepatocarcinogenesis, which have not a history of in earlier reports (11, 22). Investigating diverse polymorphisms of miRNAs and their target genes will be a serious step towards the clinical utilization of this new subclass of genetic variations in liver transplant recipients due to HCC in a population (34). Since the expression of miRNA molecules might be affected by gene polymorphisms, our objective was to explore the effect of miRNA gene polymorphisms on the outcome of liver transplantation.
MiRNA-146a is an important regulatory molecule. Studies have shown its critical role in many biological processes. MiRNA-146a can exert its proapoptotic role by blocking NF-κB, which renders to stop its effects on cell proliferation, angiogenesis, metastasis, and cancer cell survival (35, 36). Up and downregulation of miRNA-146a has been found during the activation of the innate immune system, inflammatory diseases, and cancers (37). It is believed that the dysfunction of miRNA-146a after genetic polymorphisms facilitates cancer cell migration (35, 36).
In the present study, despite the lower frequency of the CC genotype and C allele than that of the GG genotype and G allele, the CC genotype and C allele of miRNA-146a (rs2910164) polymorphisms were significantly related to the increased risk of liver rejection in HCC transplant patients. In addition, the CC genotype and C allele were significantly more frequent in male patients who experienced acute rejection. Xu et al. (38) proved in a Chinese population that the GG genotype of miRNA-146a (rs2910164) was almost two times more than the CC genotype in male participants susceptible to HCC. Higher frequencies of GG genotypes and G alleles were also found in Turkish and Korean populations (18, 24). In this study, the distribution of miRNA-146a (rs2910164) genotype GG was not different between HCC cases and controls. No significant association was found between the risk of HCC and miRNA-146a (rs2910164) polymorphism in all statistical analyses. In 2010, a study on the miRNA-146a (rs2910164) polymorphism showed that the GG genotype led to have an increased level of the miRNA molecule, causing more susceptibility to HCC (5). Another report explained that the GG genotype displayed higher levels of miRNA-146a (rs2910164) than the CC genotype (17). Another support for the recent hypothesis was produced by studying the potential targets of miRNA-146a called tumor necrosis factor receptor-associated factor 6 and interleukin-1 receptor-associated kinase 1 as critical mediators in cell growth and immune recognition (39).
In a meta-analysis, Wang et al. (37) showed that the association between miRNA-146a (rs2910164) polymorphism and susceptibility to cancer was modified by ethnicity, as well as several environmental and other risk factors that influence the population. They found the reduced effects of the miRNA-146a (rs2910164) polymorphism in Caucasians, but a borderline effect in Asian populations. Another report showed that the frequency of allele G in the miRNA-146a (rs2910164) polymorphism among different ethnicities was as follows: 0.367 in Japanese, 0.556 in Chinese, 0.763 in Caucasians, and 0.500 in Africans. This study showed that the G allele frequency of miRNA-146a (rs2910164) polymorphism was 0.752 among Iranian normal controls, similar to the previously reported allele frequency in Caucasians (International HapMap Project) (38).
The other molecule studied in the current project was. Changes in the miRNA-196a-2 expression pattern significantly affect its target function (40, 41). Also, studies revealed that a high expression level of the mentioned microRNA can facilitate cancer cell migration and invasion (41-43).
In the present study, the data showed that miRNA-196a-2 (rs11614913) genotype and allele polymorphisms had no significant effect on the transplant outcomes in liver recipients with HCC. Akkiz et al. (22) in a Turkish population reported that the CC genotype of the miRNA-196a-2 (rs11614913) polymorphism was associated with a significantly increased risk of HBV-related HCC cancer, as well as the risk of HCC in male individuals compared to women. Qi et al. (20) and Li et al. (19) also reported that the miRNA-196a-2 (rs11614913) polymorphism was related to the susceptibility to HBV-related HCC in a male Chinese population. Another study in a Chinese population reported that the miRNA-196a-2 (rs11614913) polymorphism was associated with HCC risk, and the TT genotype and T allele significantly reduced the risk of HCC when compared to the CC genotype (21). Farokhizadeh et al. (30) also reported the frequencies of CT and CC genotypes and C allele of miRNA-196a-2 (rs11614913) polymorphism were higher in HBV-positive HCC patients than in controls. However, Kim et al. (18) reported that the miRNA-196a-2 (rs11614913) polymorphism was not associated with HCC patients in a Korean population. The controversy in the genotype distribution of miRNA-196a-2 (rs11614913) and the risk of cancer might be related to ethnicity. In a meta-analysis, a significant association was found between the miRNA-196a-2 (rs11614913) TT genotype and a decreased risk of cancer in Asians compared to a Caucasian population (44).
Our results showed a relationship between the risk of HCC and polymorphisms in miRNA-196a-2, which is in agreement with other laboratory and clinical studies. As the targets of miRNA-196a-2, the HOX proteins are known for their crucial roles during embryogenesis, organogenesis, and oncogenesis (40, 45, 46). Therefore, alterations in HOX gene expression is critical in the carcinogenesis and malignant progression of HCC (47). Also, the other target of miRNA-196a-2, ANXA1, is important in transformations leading to HCC, and it is closely related to the histological grade and metastatic ability of HCC. In addition, it was shown that the up-regulation of miRNA-196a-2 could facilitate cancer cell migration and invasion (41). Changes in the expression patterns of miRNA-196a-2 can influence its targets, which might play a role in HCC susceptibility and function as an oncogenic miRNA (22). Consequently, it is sensible to propose that individuals carrying the miRNA-196a-2 (rs11614913) C allele and CC genotype might be susceptible to HCC (41).
The genetic polymorphism of miRNA-149 (rs2292832) was also evaluated in liver transplant recipients with HCC. The miRNA-149 (rs2292832) polymorphism locates in the stem-loop of mature miRNAs regions and by targeting Akt1 and E2F1 might inhibit proliferation and induce cell cycle arrest (48). Thus, alterations in the miRNA-149 gene might contribute to transplant rejection (49).
In the present study on an Iranian population, the data showed that the effects of genotypes and alleles of the miRNA-149 (rs2292832) polymorphism were not significant on HCC outcomes in liver transplant recipients. These findings are similar to those reported by Akkiz et al. (28) in a Turkish population, and He et al. (50) in a Chinese population. The mentioned polymorphism was not significantly different between HCC patients and controls but also was not similar to polymorphism reported by Kim et al. in a Korean population (18), which reported that the miRNA-149 (rs2292832) CC genotype was associated with HBV-related HCC. Also, Wang et al. in a Chinese population reported that the genotypes of miRNA-149 (rs2292832) were associated with the increased HCC risk (23).
The miRNA-499 (rs3746444) located in 3’ mature miRNAs regions might influence both the binding of 3’ mature miRNAs to Sox6 and Rod1 target genes and the cell cycle (51, 52). In the present study, the data showed that the genotypes and alleles of the miRNA-499 (rs3746444) polymorphism had no significant effect on the HCC outcomes in liver transplant recipients. The findings of our study are in line with those reported by Akkiz et al. (24) and Wang et al. (23) that found no significant association between miRNA-499 (rs3746444) and susceptibility to HCC. However, our findings are in contrast to those reported by Kim et al. (18) that found the miRNA-499 (rs3746444) AA genotype was significantly associated with the risk of HCC and Shan et al. (53) that found the miRNA-499 (rs3746444) AG genotype was significantly associated with the reduced risk of HCC. The miRNA-499 (rs3746444) G allele was observed as a risk factor for cancer in an Asian population (54). The importance of ethnicity was also reported. The results of this study re-emphasize the importance of human genetic SNPs on the initiating and complicating clinical outcomes in transplant recipients, as presented in earlier reports (55-57). However, this study suffered limitations about the size and the type of samples that should be considered in the evaluation of results. We need further completed studies to validate these findings. The study was retrospective, and explanted liver of all recipient candidates with HCC who admitted for transplant were included. The collected samples were stored in a sample bank and we had no access to blood samples of liver recipients and donors and liver tissue of donors to evaluate.
5.1. Conclusions
The results showed that the CC genotype and C allele of the miRNA-146a (rs2910164) polymorphism are significantly associated with a higher risk of graft rejection in liver transplant patients with HCC. Further independent studies are required to validate these findings in a larger population and in patients with different ethnicities.