1. Background
Hepatitis C virus (HCV) belongs to the Flaviviridae family, characterized by its small size, enveloped appearance, and 9.6 kb positive-sense single-stranded RNA. HCV encodes structural proteins, including highly glycosylated enveloped E1, E2, and Core (C), and also non-structural proteins (NS), including NS2, NS3, NS4A, NS4B, NS5A, NS5B, and P7. Structural proteins are important for both HCV life-cycle and its entry to the host cell. While non-structural proteins are essential for viral RNA replication and virus morphogenesis (1, 2).
HCV causes both acute and chronic infections. In most cases, acute infection is asymptomatic and rarely can be diagnosed. Moreover, in those who are developing chronic HCV, the infection is also undiagnosed in most cases, until developing symptoms such as liver diseases (cirrhosis and hepatocellular carcinoma) (1, 3). Globally, nearly 71 million people (1% of the world’s population) are living with chronic hepatitis C virus infection (4). In most cases (50% - 90%), HCV is asymptomatic. Only 20% - 30% of infected patients are aware of their disease (5). HCV RNA virus transmits primarily via the blood route, including blood transfusion, unsafe injection, etc. (6). HCV represents a major cause of morbidity and mortality among people who receive hemodialysis (HD) (7). According to our previous report conducted in Iran (2016), the incidence of HCV seroconversion among chronic HD patients is about 3.36% (8).
Various virological tests have been developed based on the specific identified part of HCV virus. At present, HCV diagnosis begins with screening enzyme immunoassay (EIA) (9).
The first generation of HCV ELISAs used recombinant proteins complementary to the non-structural protein 4 (NS4) as antigen. The window period (WP) of the first generation, the time between infecting with HCV and the appearance of detectable antibodies, was 4 - 6 months (9). The second generation assay had higher sensitivity and specificity than the first and was based on NS3, NS4, and core as antigens (10). In 1996, the sensitivity of the test was further improved in the third-generation, the main difference was the addition of NS5 to those used in the second-generation. The third-generation test has successfully reduced the window period to 7 - 8 weeks (11, 12).
Eventually, in 2003, the fourth-generation assay or antigen–antibody combo assay was introduced. This generation is based on Core, NS3, NS4A, NS4B, and NS5A antigens of HCV genotypes 1a and 1b along with NS3 and NS4 antigens from HCV genotypes 2 and 3a, in combination with antibodies against Core antigen for antigen detection. Therefore, fourth-generation assays detect simultaneously antibodies for genotypes 1a and 1b as well as 2 and 3a (13, 14). Furthermore, because of its high sensitivity, the WP is decreased to 26 days (9). ELISA tests are both reliable and cost-effective. Therefore, they can be applied widely as the first-level screening procedure. Although it has many advantages, but the results of ELISA based assays should be confirmed by a recombinant immunoblot assay (RIBA), that is expensive (15). However, third-generation ELISAs, the mostly used detection kits, cannot be used before the seroconversion step, and because of high false-positive results, its positive predictive value (25%) in low-risk people is often low (16, 17). Because end-stage renal disease (ESRD) has a long WP and these patients are at increased risk of acquiring HCV through HD, finding a reliable, accurate and sensitive diagnostic test for HCV infection is of crucial importance (18). In addition, third and fourth generations of ELISA kits are commonly based on an indirect ELISA format, which is less sensitive than antigen/antibody Sandwich ELISA (19, 20). In this line, the current study aimed to generate a more reliable and sensitive diagnostic tool for HCV infection.
2. Objectives
To achieve the study goal, an ELISA based-platform was developed, which is designed to detect E2 protein as an antigen with other serum antibodies against core, NS3, NS4, and NS5 antigens. As mentioned earlier, E2 is a viral structural protein, which contains the viral membrane. Unlike core protein, E2 protein could be detected without heating the serum samples. In this study, the antigen sandwich ELISA was used to improve the sensitivity of the test. This technique is based on indirect methods, contrary to the third generation ELISA which uses direct methods. Moreover, we used biotinylated second antibodies or antigens to amplify signals and increase the detection sensitivity.
3. Methods
3.1. Subjects and Serums
Serum specimens were collected from volunteer whole blood donors in the Iran blood transfusion organization (IBTO) and patients who were receiving hemodialysis treatment in hemodialysis centers of Tehran and Bandar Abbas cities.
In total, 107 positive and 415 non-reactive for antibody test samples from IBT. Since most of the blood donors are healthy, random sampling was limited and confirmed positive and non-reactive samples were collected. Positive samples were confirmed using the NAT. For samples collected from HD patients, 202 and 2 were negative and positive for the HCV NAT test, respectively.
Since the negative predictive value (NPV) of IBTO’s standard screening tests is high, negative results for HCV were considered as negative, but the positives re-tested using the NAT, which is the gold standard for evaluating active HCV infection (6). Samples from IBTO were used as approved specimens to evaluate the sensitivity and the specificity of the current methods. Furthermore, HD patients were considered as antibody production deficient with high false-negative results in antibody detection serologic assays. Approved results of the IBTO were considered as inclusion criteria for blood donor samples. For HD patients, the quality of the samples was used as inclusion criteria. Hence, samples with high hemolysis and lipemic and icteric indexes were excluded. Written informed consent was obtained from all participants. Moreover, the addiction status of all participants, blood transfusion, jaundice, or liver disease were evaluated.
3.2. Materials
Monoclonal antibody of HCV envelope glycoprotein E2 (Capturing antibody for antigen detection), mouse anti hepatitis C E2 antigen (detective antibody for antigen detection), Rabbit anti-human IgG antibody, and Rabbit Anti-Human (IgM antibody) were purchased from Acris antibodies, AbD serotec, thermo fisher scientific, and Abcam companies, respectively. Escherichia coli-derived NS3 (genotype 1a, amino acids 1192 - 1459), NS4 (Mosaic amino acids, 1691 - 1710, 1712 - 1733, 1921 - 1940 from genotypes 1, 2, 3, and 5), NS5 (genotype 1b, amino acids 2061 - 2302), and Core24 (genotype 1b, amino acids 2 - 119) antigens were purchased from RPC Company. Additionally, streptavidin-peroxidase (HRP) (Cat# 11089153001) and N-hydroxysuccinimide (NHS) ester-biotin (Cat# PG82075) were purchased from Roche and PierceTM. All buffers and solutions were prepared with ultrapure water.
3.3. Bioconjugations
Biotinylated secondary antibodies and antigens were used to amplify the detection signal of proteins expressed at low levels (21). For Biotinylating, a stock solution of biotin-NHS in DMSO (dimethyl sulfoxide) (Sigma, USA) was prepared at 40 mg/mL concentration. Next, 6 μL stock biotin-NHS was added to 2 µL antibody solution (0.5 mg/mL), following by 2-hour of stirring in a dark container at room temperature. 6 µL of 0.5 mol/L ethanolamine was added to the solution, incubated for 30 minutes. The biotin conjugation of antigens was performed using the aforementioned protocol with some modifications to optimize the concentrations of protein and conjugation reagent.
3.4. Tests Set up Procedure
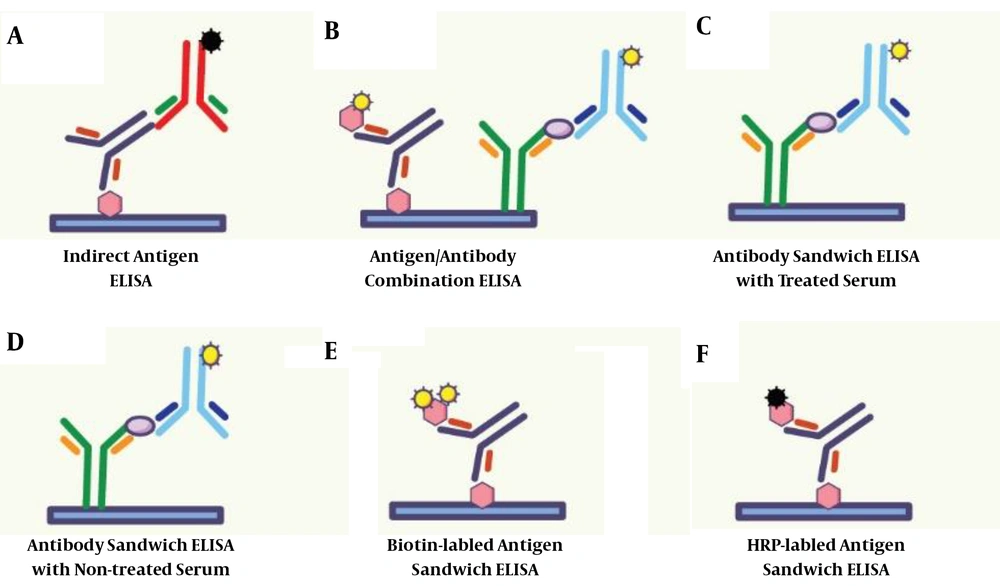
In this research, six methods were used for ELISA tests, including indirect ELISA, antigen sandwich ELISA, biotinylated antigen sandwich ELISA, Anti-E2 Antibody Heat-Treated sandwich ELISA, Anti-E2 antibody not-treated sandwich ELISA, and antigen/antibody combination ELISA. These six experiments were used to test HCV in all samples. In Indirect ELISA, antigen sandwich ELISA, and biotinylated antigen sandwich ELISA, one type of Escherichia coli-derived NS3, NS4, NS5, and Core24 antigen were immobilized on flat-bottom polystyrene ELISA plate. Simultaneously, in Anti-E2 antibody heat-treated/non-treated sandwich ELISA, anti-E2 antibodies were immobilized on the ELISA plate. Moreover, in antigen/antibody combination ELISA, both HCV antigens and anti-E2 antibodies were used to immobilize on the ELISA plate. To generate a detection signal in the mentioned ELISA tests, HRP conjugated rabbit anti-human IgG/IgM polyclonal antibodies, HRP conjugated antigens, and Biotin-labeled antigen/anti-E2 antibody were used. These processes are described in Figure 1.
The coating and the blocking buffer solutions, temperature, time, sample volume, the concentration of the coating antigen, streptavidin-HRP and biotin dilution ratio, and the reaction time were optimized and determined to set up an appropriate strategy for the ELISA test.
Antigens or antibodies were coated on the polystyrene flat-bottom ELISA plate (96 well MicroWellTM MaxiSorpTM flat bottom plate) according to the methodology used in our previous study (22). After coating the ELISA plates with antigens and HCV/anti-E2 antibody, PBS/ bovine serum albumin (BSA)-2% (Gilden west)/threhalose-3%/gelatin-1.5% was used as the blocker for 1hour at room temperature. Then, 200 µL of the sample diluent and specific volume of samples, positive and negative controls (10 µL for indirect ELISA and 50 µL for other ELISA tests) were added to each well. The plate was incubated for 1 h at 37°C, after which, PBS/Tween 20% - 0.02% was used to wash the plates (five times). Subsequently, mouse polyclonal anti-human immunoglobulin in indirect ELISA and HRP/biotin-labeled HCV antigens in antigen sandwich ELISA were added for recognizing serum anti-HCV antibodies. In biotin-labeled assays, the streptavidin-HRP conjugate was used as a signal producer conjugate in combination with the biotinylated conjugate reagent. In the heat-treated and non-treated antibody sandwich ELISA method, the biotin-labeled antibody was utilized. Then, washed for five times using the wash buffer to remove unbound conjugate. Afterward, 100 µL of Tetramethylbenzidine (TMB) substrate was added and left at room temperature for 15 minutes. After stopping the reaction, plates were read at 450 nm, regarding 630 nm on an ELISA Reader (BioTek). To determine the cut-off value, the mean and standard deviation of negative specimens were used.
For samples with different results in each HCV ELISA test, real-time PCR viral load testing was used. Serum HCV RNA levels were measured using the COBAS AmpliPrep/COBAS TaqMan HCV test, which includes automated sample preparation on the COBAS AmpliPrep followed by real-time PCR and COBAS TaqMan detection in accordance with the manufacturer’s instructions.
3.5. Statistical Analysis
To calculate the sensitivity, specificity, accuracy, and positive and negative predictive values of each assay, standard equations were used. Variables were compared in a pairwise fashion using McNemar’s, westgard QC recommendation, and Medcalc Software Ltd, tests. Statistical analyses were performed by SPSS version 21 (Windows).
4. Results
4.1. Demographic Characteristics
In this study, 726 samples were analyzed for HCV. 49 and 51% of samples were obtained from females and males, respectively, and the mean age of participants was 38.9 ± 16.97 years, ranging from 18 to 87 years.
4.2. Comparisons Among Different ELISA Methods in HCV Diagnosis
All collected samples (726) were categorized into five groups as follows: Group A, included 415 negative blood donor samples; group B, included 107 positive blood donor samples confirmed by NAT method; group C, included 202 samples of HD patients’ with negative HCV results according to both third-generation ELISA test and RT-PCR; group D, included one HCV positive patients with HD that was positive in both third-generation ELISA test and RT-PCR; and group E, included one HD patient who was positive only by real-time PCR.
All discrepancies were addressed by re-testing, whether for false positive or false negative tests.
Antigen sandwich (HRP) ELISA test was presented as follows: Group A, 414 negative samples and 1 positive sample; group B, 104 positive samples and 3 negative samples; group C, 201 negative samples and 1 positive sample; group D, one positive; and group E one sample was negative.
The results of antigen sandwich (Biotin) ELISA were as follows: Group A, 414 negative and one positive sample; group B, one negative and 106 positive samples; group C, 201 negative and one positive sample; group D, one positive sample; and group E, one negative sample.
Analyzing samples with combined antigen and antibody ELISA indicated: Group A, 1 positive sample and 414 negative samples; group B, totally positive samples; group C, 201 negative samples and 1 positive sample. All samples of group D and E were positive.
The findings of the E2 antigen sandwich ELISA revealed no difference between treated and non-treated results, as follows: Group A, all samples were negative; group B, 104 positive and three negative samples; group C, all samples were negative; and groups D and E, All samples were positive.
In the present study, the combined Ag/Ab test had the highest accuracy and sensitivity as well as the lowest false positive and negative results. It can be attributed to the fact that combined Ag-Ab assay has the highest accuracy and sensitivity. Besides, it worth noting that the antibody sandwich assay has the highest specificity.
The results of all tests separated by six assays are described in Table 1. As mentioned above, the samples were evaluated using various assays. Also, a comparison of all six assays is provided in Table 2.
| Comparative Method | Real-Time (Viral Load) | |||||
|---|---|---|---|---|---|---|
| Test Method | PPA% (CI) | PNA% (CI) | POA% (CI) | Diagnostic Sensitivity% (CI) | Diagnostic Specificity% (CI) | Diagnostic Accuracy (CI) |
| Indirect | 95.4 (89.7 - 98) | 99.2 (98.1 - 99.7) | 98.6 (97.5 - 99.3) | 95.41 (89.62 - 98.49) | 99.19 (98.12 - 99.74 | 98.62% (97.48 - 99.34) |
| Ag sandwich (HRP) | 97.2 (92.2 - 99.1) | 99.7 (98.8 - 99.1) | 99.3 (98.4 - 99.7) | 97.22 (92.10 - 99.42) | 99.68 (98.84 - 99.96) | 99.31 (98.4 - 99.67) |
| Ag sandwich (biotin) | 98.2 (93.6 - 99.5) | 99.7 (98.8 - 99.9) | 99.4 (98.6 - 99.8) | 98.17 (93.53 - 99.78) | 99.68 (98.83 - 99.96) | 99.45 (98.60 - 99.85) |
| Ab sandwich (treated) | 98.2 (93.6 - 99.5) | 100 (99.4 - 100) | 99.7 (99.0 - 99.9) | 98.17 (93.53 - 99.78) | 100 (99.4 - 100.00) | 99.72 (99.01 - 99.97) |
| Ab sandwich (non-treated) | 98.2 (93.6 - 99.5) | 100 (99.4 - 100) | 99.7 (99.0 - 99.9) | 98.17 (93.53 - 99.78) | 100 (99.4 - 100.00) | 99.72 (99.01 - 99.97) |
| combined Ag and Ab | 100 (96.6 - 100.0) | 99.7 (98.8 - 99.9) | 99.7 (99.0 - 99.9) | 100 (96.67 - 100) | 99.6 (98.83 - 99.96) | 99.72 (99.01 - 99.97 |
Abbreviation: PNA, predictive negative agreement; POA, predictive overall agreement; PPA, predictive positive agreement.
| Test Type | Exact Sig. (2-tailed) |
|---|---|
| Indirect-Ag sandwich (HRP) | 0.625 |
| Indirect-Ag sandwich (biotin) | 1.000 |
| Indirect-combined Ag and Ab | 0.727 |
| Indirect-real-time | 1.000 |
| Ag sandwich (HRP)-Ag sandwich (biotin) | 0.500 |
| Ag sandwich (HRP)-combined Ag and Ab | 0.125 |
| Ag sandwich (HRP)-real-time | 0.687 |
| Ag sandwich (biotin)-real-time | 1.000 |
| Ab-sandwich-non-treated and combined-Ag-Ab | 0.063 |
| Combined Ag and Ab-real-time | 0.5 |
| Real-time-and Ab-sandwich-treated | 0.250 |
| Real-time- and Ab-sandwich-non-Treated | 0.250 |
| Ab-sandwich-treated and combined-Ag-Ab | 0.063 |
Calculations of PPA, PNA, and POA were done by 2 × 2 Contingency Calculator: westgard QC Copyright© 2020. All rights reserved. Westgard QC, Inc., 7614 Gray Fox Trail, Madison WI 53717Call 608-833-47183 or e-mail us at westgard@westgard.com available online at http://tools.westgard.com/two-by-two-contingency.shtml
Calculations related to diagnostic sensitivity, diagnostic specificity, and diagnostic accuracy were performed by Medcalc (easy to use statistical software) ©2020 MedCalc Software Ltd. Available online at https://www.medcalc.org/calc/diagnostic_test.php.
The value of sample‐to‐cut off (s/co) ratio in various methods was evaluated, and the obtained median of the results was 3.75 for indirect, 4.7 for Ag sandwich (HRP), 6.5 for Ag sandwich (biotin), 4.6 for Ab sandwich (treated), 4.2 for Ab sandwich (non-treated) and 8.4 for combined Ag and Ab ELISA methods.
5. Discussion
HCV causes both acute and chronic infections. Acute HCV usually is not a life-threatening disease. However, those with chronic HCV are at increased risk of complications such as cirrhosis. Early diagnosis is of crucial importance to prevent the development of chronic HCV infection (23). Recent studies have estimated a low prevalence of HCV in Iran (24). However, its incidence is on the rise through shared injection. Besides, its prevalence is significantly high among HD patients, ranging from 7.6 to 13.6% (ELISA (13.6%), RIBA (12.2%), and PCR (7.6%)). Therefore, early detection and management among HD patients should have a high priority due to the high risk of HCV transmission through hemodialysis devices (25, 26).
The serological measurement of the anti‐hepatitis C virus antibody is widely using as the first-line diagnosis of HCV infection, and, therefore, increasing the performance of this criterion is of crucial importance for HCV screening. Currently, four generations of ELISAs are available to detect HCV infection. The third-generation ELISAs are the mostly used detection kits. These kits recognize antibodies against virus antigens, including NS3, NS4, NS5, and Core via indirect method. In other words, third-generation kits cannot detect the infection before seroconversion. Besides, they also have high false-positive results when performing on low-risk individuals and immunocompromised patients (27). In this study, antigen sandwich ELISA was used, which presented more accurate, sensitive, and specific results than indirect ELISA methods. In fourth-generation ELISA, in addition to antibodies that can be recognized by the third-generation kits, it is possible to identify Core antigens. However, to decrease the destruction of serum antibodies, samples should not be heated, which limits the detection of Core antigen by the fourth-generation kits. In this study, instead of Core antigens, E2 protein, which is located on the HCV surface, were detected. Hence, we could detect E2 protein without heating the serum samples. Incidentally, the results showed that heated samples are less accurate than not heated ones.
In conclusion, in this study, the sensitivity, specificity, and accuracy of the combined antigen and antibody assays were higher than the indirect and antigen sandwich assay. The sensitivity, specificity, and accuracy of the combined Ag/Ab ELISA test were similar to the real-time PCR results. Also, the Ag/Ab ELISA test showed high signal/noise ratios. This test could be used as a screening assay for HCV detection, based on its high accuracy, sensitivity, and specificity. The current study had limitations, including not considering the effects of tests on the WP. Besides, the results of seroconversion patients should be evaluated in future studies to determine the WP in Ag ELISA and Ag/Ab ELISA tests. Besides, the structure of E2 proteins is more variable than Core proteins. Therefore it is difficult to design a monoclonal antibody for them to detect all different E2 genotypes. The authors recommend comparing the results of this method with fourth-generation kits in future studies. Also, samples from Auto cleared patients, patients with rapid virological response (RVR), and sustained virological response (SVR) should be tested by this novel method to investigate the correlation of the obtained result with the results of evaluating viral load via NAT assay. Moreover, it should be determined whether this ELISA method can be used as a quantitative test to measure viral load or not, like what we see in Real-time PCR.