1. Background
Hepatitis C Virus (HCV), a positive-sense single-strand RNA virus, entails a serious and growing public health problem, chronically infecting about 70 million people worldwide. In most cases, this blood-borne pathogen can establish a chronic infection, end-stage liver disease, cirrhosis, and hepatocellular carcinoma (1, 2).
Although viral replication occurs primarily in the liver, HCV can infect and replicate into most human cells and tissues, causing local and systemic inflammation. Consequently, HCV infection is considered a systemic disease. In recent years, Direct-acting Antiviral Agents (DAAs) have been approved for treating HCV. These drugs can achieve HCV clearance in up to 98% of cases with a favorable clinical impact on hepatic and extrahepatic manifestations (3, 4). Yet, an estimated 1.5 - 2 million new HCV infections occur globally each year (5). A Sustained Virologic Response (SVR) occurs when blood tests continue to show no detectable RNA 12 weeks or longer after treatment.
The HCV is no longer considered to be only hepatotropic because the HCV RNA has been detected in extrahepatic sites, such as Peripheral Blood Mononuclear Cells (PBMCs) (6, 7), bone marrow (8), and the central nervous system (9). Studies have shown that in some cases, the HCV genotype in plasma is different from that in PBMCs (10, 11). The main prognostic factor for HCV infection treatment is a genotype of HCV RNA in extrahepatic sites. After the cure, at least with interferons, PBMCs are a popular site for HCV replication (12). However, the goal of hepatitis C therapy is to eliminate the virus from the liver tissue or peripheral blood mononuclear cells. It is argued that after liver transplantation, HCV replication in extrahepatic sites is the primary cause of reinfection or relapse of HCV infection (13). Some reports show that HCV RNA can be detected in peripheral mononuclear cells of patients with chronic hepatitis C virus infection and, thus, its possible contribution to the pathogenesis of the extrahepatic disease. Besides, PBMCs may be the preferred sites for virus replication and virologic relapse after successful antiviral therapy, even if SVR occurs (13-16).
There is some controversy about whether the HCV can only bind to the mononuclear cells' cellular receptors, or the virus can get into the cell and replicate in those cells (17). Studies have shown that HCV can bind to low-density lipoprotein (LDL) and enter PBMCs via LDL-receptors (18-20). It is widely understood that CD81 (a molecule in the tetraspanin superfamily) has a significant key role in the HCV entry process (21). Even if the HCV test is negative, small numbers of replicating viruses in PBMCs may cause the infection to return.
2. Objectives
This study examined if HCV can replicate in vitro inside mononuclear cells isolated from infected patients with and without SVR. The presence of replicative forms of RNA in PBMCs indicates an extrahepatic site for HCV replication.
3. Methods
3.1. Patients and Samples
Sixteen patients with chronic HCV infection were selected from patients referring to the Gastroenterology and Hepatology Clinic, Shiraz University of Medical Sciences, from July 2018 to October 2018. The patients were 13 males and three females, and the age range was 28 to 50 years. Participants gave written informed consent, and a local ethics committee approved the study.
Based on the clinical data collected at the beginning of the study, all patients were positive for the anti-HCV antibody in plasma for more than six months before starting treatment. The HCV RNA in plasma was positive before beginning treatment. All patients were on pegylated interferon alpha 2a (peg IFNα2a) and ribavirin treatment. Four of the patients showed initial SVR at week 12 post-treatment. The remaining patients were positive for HCV RNA on the sample collection day. The exclusion criteria included HBV, HIV, or other chronic diseases and the history of previous treatment with antiviral drugs.
Blood samples were collected from patients who were found to have a positive PCR test for HCV RNA in plasma samples, and the patients achieved SVR at week 12 post-treatment. From each participant, 5 mL of blood was collected into a sterile EDTA-containing vacutainer tube. Plasma was separated by centrifugation and stored at −80°C for future investigation. Then, PBMCs were isolated from whole blood by Ficoll-Hypaque density gradient centrifugation, and then the RPMI 1640 medium was used for re-suspending the pellet.
3.2. Lymphocyte Culture
For the detection of the replicative form of HCV RNA in PBMNs, between 106 - 107 viable cells isolated from all the patients' blood samples were cultured in the RPMI 1640 medium and stimulated with optimized concentrations of IL-2 (10 mg/ml) and PHA (5 mg/ml) (22). The viability assay and cell count were carried out on the day of sample collection and one, three, and seven days after lymphocyte culture.
3.3. Extraction of Viral RNA from Plasma and PBMC Specimens and cDNA Synthesis
The viral RNA from plasma samples and lymphocyte culture on days one, three, and seven (1×106 cell/mL) were extracted using the TRIzol® extraction Kit (Life Technologies, Rockville, MD) following the manufacturer's instruction. The cDNA was synthesized using the First-strand Synthesis Kit with a random hexamer primer (Fermentas, Lithuania).
3.4. Detection and Quantification of Negative-Strand Hepatitis C Virus RNA in PBMNCs
For the detection and quantification of HCV negative-strand, real-time reverse transcriptase-PCR (RT-PCR) was performed using specific primers for the amplification of 5' noncoding (5'NCR) of the HCV genome. The primers' specificity against the target sequence was determined using the Primer-BLAST program and the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/tools/primer-blast/).
The real-time PCR was carried out after a reverse transcription step. For the quantitative detection of negative-strands of HCV RNA, the SYBR Green method was carried out for each sample with a set of primers for 5'NCR. The forward primer 5' GGCGACACTCCACCATAGATC 3' and the reverse primer 5' GGTGCACGGTCTACGAGACCT 3' were designed to amplify a 324 bp fragment within the 5' noncoding region of the HCV genome (positions 1 - 324). Quantitation was carried out using an external standard curve. The standard curves for the quantification of HCV RNA 5'NCR were constructed using the method previously described (2).
3.5. Real-Time PCR Assay
Real-time PCR was performed with Thermo Scientific Maxima SYBR Green qPCR Master Mix (2x) kit (Thermo Scientific, Germany) following the manufacturer's instructions. The thermal cycling program was designed as follows: initial denaturation at 95°C for 10 min, followed by 40 amplification cycles consisting of initial denaturation at 94°C for 40 s, annealing at 59°C for 30 s, and extension at 72°C for 30 s. At the end of each PCR run, the system automatically analyzed data, and amplification plots were generated.
3.6. Genotyping
To determine HCV genotypes, RNA extracted from plasma and lymphocyte cultures was tested using Versant HCV Genotype 2.0 assay (INNO-LiPA HCV 2.0) following the manufacturer's instructions. For the patients with SVR, the HCV genotyping test results were obtained by reviewing their medical files with the permission of their physicians.
3.7. Statistical Analysis
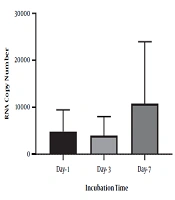
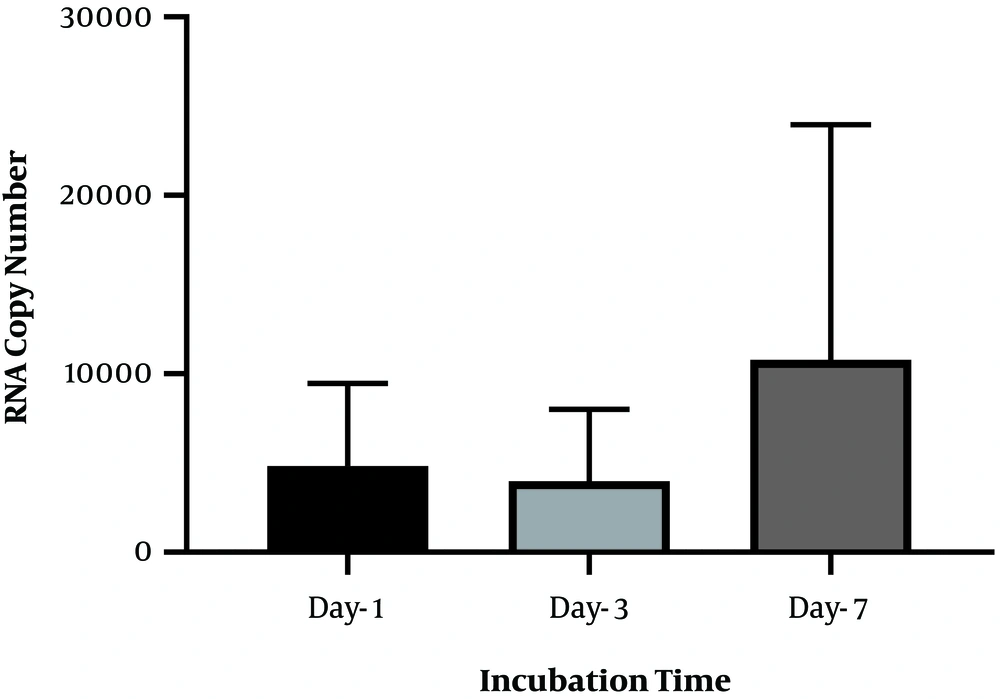
Statistical analysis was performed using GraphPad Prism version 7.01 software for Windows (Graph Pad Software, San Diego, CA, USA). The one-way ANOVA and Tukey's multiple comparison tests were used to compare the results obtained from quantitative real-time PCR in three different days after lymphocyte culture.
4. Results
4.1. Mononuclear Cell Count in Culture Medium at Different Incubation Times
The cell count was done in different incubation periods (collection days, day three and day seven) at the optimal concentrations (5 μg/mL) of PHA mitogen and IL-2 (10 μg/mL) (Table 1). The results showed that seven days’ incubation time was optimum for mitogen to obtain the maximum number of viable cells (P-value < 0.05).
| Sample | PBMCs, cells/mL | ||
|---|---|---|---|
| Day 1 | Day 3 | Day 7 | |
| 1 | 1.8 × 106 | 1.38 × 106 | 3.36 × 106 |
| 2 | 2.1 × 106 | 1.76 × 106 | 5.6 × 106 |
| 3 | 1.84 × 106 | 2.96 × 106 | 3.76 × 106 |
| 4 | 1.14 × 106 | 1.88 × 106 | 2.92 × 106 |
| 5 | 1.8 × 106 | 1.88 × 106 | 2.8 × 106 |
| 6 | 8.5 × 105 | 1.2 × 106 | 3.08 × 106 |
| 7 | 6 × 106 | 1 × 106 | 2 × 106 |
| 8 | 9 × 106 | 1.28 × 106 | 3.2 × 106 |
| 9 | 1.38 × 106 | 1.6 × 106 | 3.6 × 106 |
| 10 | 1.78 × 106 | 1.4 × 106 | 3.48 × 106 |
| 11 | 1.9 × 106 | 1.08 × 106 | 2.72 × 106 |
| 12 | 1.44 × 106 | 1.2 × 106 | 2.8 × 106 |
| 13 | 1.22 × 106 | 1.04 × 106 | 2.6 × 106 |
| 14 | 9.6 × 106 | 1.44 × 106 | 3.36 × 106 |
| 15 | 1.4 × 106 | 1.12 × 106 | 4.12 × 106 |
| 16 | 2.02 × 106 | 1.92 × 106 | 4.8 × 106 |
4.2. Detection of Negative-Strand HCV in Mononuclear Cells Over Time by Real-Time PCR
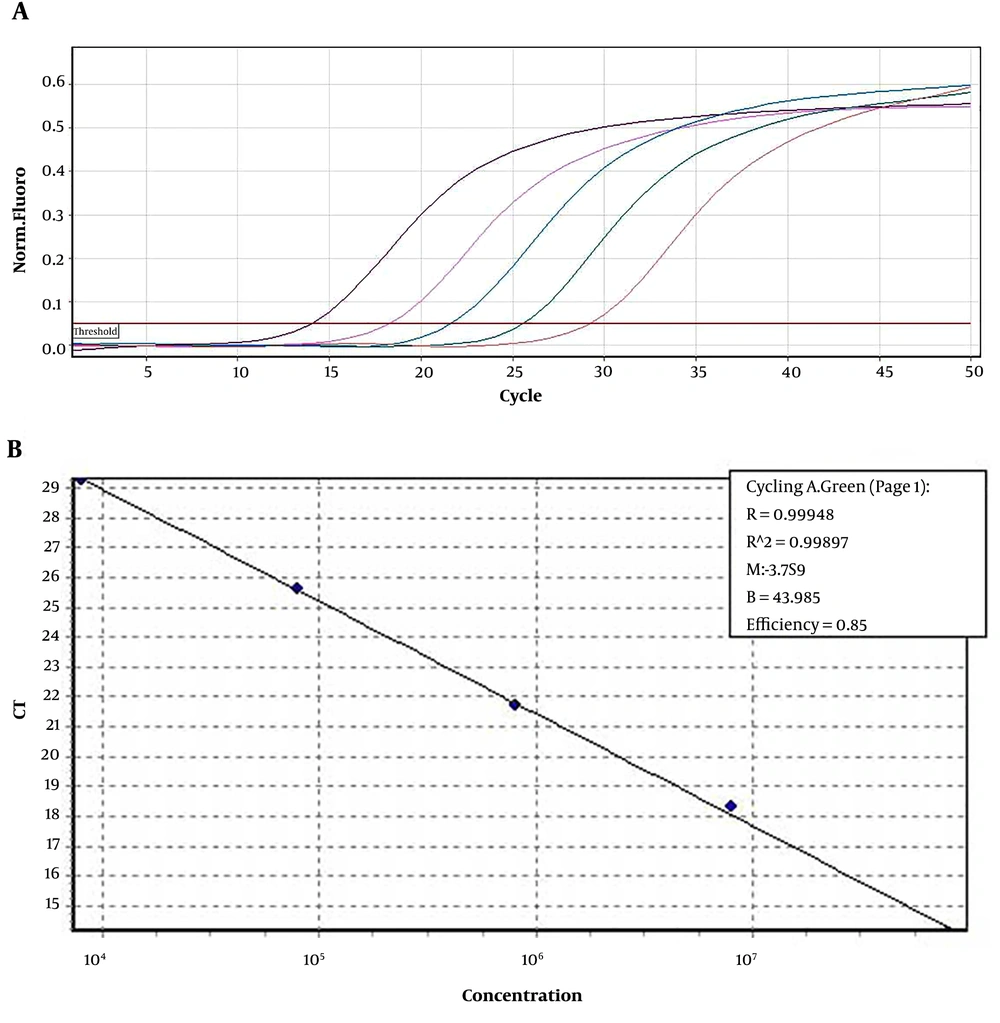
The Limit of Detection (LOD) of real-time RT-PCR was determined as 20 RNA copies/106 mononuclear cells. The amplification plots and the standard curve are shown in Figure 1.
Using constructed plasmids containing 5'NCR coding regions and production of control RNA sequence from different areas of the HCV genome, standard curves were plotted to quantify SYBR green real-time PCR. The quantitative real-time PCR efficiency for HCV 5'-NCR was 0.85, and the R2 values for all regions were 0.99.
To identify the negative-strand HCV in mononuclear cells harboring HCV, we performed RNA extraction at different incubation times, followed by strand-specific RT-PCR, to detect the minus strand of HCV. The minus-strand was not consistently detected in mononuclear cells isolated from HCV-infected patients. The 5' NCR region of negative-strand HCV RNA was found in 10 out of 16 (62.5 %) samples (Table 2). As shown in Table 2, among four samples collected from patients with SVR, negative-strand HCV RNA was found in two patient samples by primer specific RT-PCR to detect the 5' NCR region after PBMC culture. Overall, the increasing level in the minus strand of HCV RNA was found overtime in samples that were positive for HCV RNA. In one patient with SVR, an increased level of HCV RNA was found in lymphocyte culture on day three and day seven after cell culture incubation by specific primers (patient 10). Although no significant differences were found between the number of RNA copies in three different days after cell culture (P-value = 0.1), the overall increase in the level of HCV RNA was found in some samples over time (P-value < 0.01) (Figure 2). All plasma samples were tested by real-time PCR on the collection day before cell culture (Table 2).
| Sample | Age (y) | Sex | RNA Copies/Ml in Plasma Samples | Negative-Strand HCV RNA Copies/1×106 Mononuclear Cell Culture | Genotype | ||
|---|---|---|---|---|---|---|---|
| Day 1a | Day 3 | Day 7 | |||||
| 1 | 31 | M | 160 | 991 | 941 | 856 | 1a |
| 2 | 29 | M | 821 | 1482 | 1230 | 9883 | 3a |
| 3 | 43 | M | 5420 | 4485 | 4459 | 18628 | 1a |
| 4 | 39 | M | 373 | 12915 | 730 | 716 | 3a |
| 5 | 47 | M | 257 | 155 | 261 | 745 | 1b |
| 6 | 28 | F | 10313 | 5971 | 4328 | 44687 | 3a |
| 7 | 36 | M | 24375 | 5415 | 4496 | 997 | 1a |
| 8 | 33 | M | 3099 | 1260 | 2109 | 4756 | 3a |
| 9 | 29 | M | 5118 | 13093 | 12145 | 17875 | 1a |
| 10b | 46 | M | UD | 1240 | 2225 | 5428 | Undeletable |
| 11b | 41 | F | UD | UD | UD | UD | 1a |
| 12b | 28 | M | UD | UD | UD | 72 | 3a |
| 13b | 36 | M | UD | UD | UD | UD | 1b |
| 14 | 42 | M | 2643 | UD | UD | UD | 1a |
| 15 | 36 | M | 2517 | 6531 | 10869 | 14143 | 1b |
| 16 | 30 | M | 1146 | UD | UD | 20 | 1a |
Abbreviations: SVR, Sustained virological response; M, male; F, female.
aDays after incubation.
bPatients with SVR, UD: undetectable.
5. Discussion
In the present study, we investigated post-treatment persistence and the possible replication of intracellular HCV in peripheral blood mononuclear cells of 16 patients with and without Sustained Virological Response (SVR). The quantitative and specific detection of negative-strand HCV RNA extracted from patients’ PBMN cell culture was performed over time. The real time-PCR assay's specificity and sensitivity were assessed with a serial dilution of RNA control obtained from a plasmid containing HCV cDNA (2).
The detection of HCV RNA in serum samples and PMBN cells on the collection day confirmed the existence of active HCV infection in 12 patients at week 12 post-treatment. In comparison, HCV RNA was undetectable in PBMN cells or serum samples of four patients on the same treatment protocol.
The analysis of negative-strand HCV RNA extracted from PBMN cell cultures indicated that HCV could replicate in PBMN cells of the majority of patients who were still on treatment. Nevertheless, in two patients with SVR, HCV RNA was detected in cell culture. In one patient, a low level of HCV RNA (72 copies) was found on day seven after cell culture. The results indicated possible recurrent infection from PBMN cells as extrahepatic HCV reservoirs in patients who achieved SVR.
It has been shown that HCV is not a strictly hepatotropic virus (23, 24). There is some evidence that shows the presence of HCV RNA in peripheral blood mononuclear cells as extrahepatic reservoirs; however, a few studies indicated the replication of the virus in PBMN cell culture isolated from the patients. However, detecting either sense or antisense RNA genomic strands is essential for predicting post-treatment relapse of HCV infection (6, 25, 26). All types of mononuclear peripheral mononuclear cells, including monocytes/macrophages and lymphocytes, harbor the HCV RNA (27).
In the present study, a decreased level negative-sense HCV RNA was determined in three out of 12 (25%) in vitro cell cultures over time, indicating that viral treatment inhibits the replication of the viral RNA. Nonstructural protein 5B (NS5B), an RNA-dependent RNA polymerase, plays a crucial role in replicating the HCV RNA. Effective treatment inhibits the expression of the protein and results in the prevention of RNA replication. Despite HCV RNA positivity in the serum sample of one patient, the HCV RNA was not detected in cell culture on day one and day three after incubation (patient 16). However, the lowest number of HCV RNA negative-sense was found seven days after incubation. The possible explanation is the presence of rare infected cells on the collection day, although using PHA and IL-2 may stimulate the virus replication in activated cells. From four patients who achieved SVR, a high copy number of HCV RNA was found in PBMN cells of one patient (patient 10) on day one, which increased over time. The RNA extraction and PCR assay were repeated on the same serum sample, but no positive result was obtained. A low copy number of negative-strand HCV RNA was detected in the cell culture of an SVR patient (patient 12) seven days’ post-incubation.
Considering that HCV can infect PBMN cells, we can assume that HCV replication may take place in some negative cases, but the low level of RNA within the cells is beyond the detection limit of the assay.
In conclusion, this study indicated that PBMCs can be the source of recurrent HCV hepatitis infection after SVR and antiviral treatment. Viral genotype did not play an essential role in virus replication in vitro. It is crucial to investigate if the viral protein is expressing within the cells, as well. Electron microscopy will support the presence of viral particles within the cells if any.