1. Context
Hepatitis C virus (HCV) infection is one of the major public health concerns and a major cause of chronic cirrhosis and hepatocellular carcinoma (HCC) in the world (1). Approximately 30% of infected patients spontaneously clear the virus from their bodies, although the majority of the patients develop a chronic infection that can lead to cirrhosis and HCC (2).
Interaction between HCV and immune responses of the host plays an important role in the outcome of HCV infection, although the precise mechanisms underlying the spontaneous viral clearance or development of chronic HCV infection are not fully understood (3). Human leukocyte antigen class-II (HLA-II) is believed to play an important role in immune responses by presenting HCV antigens to CD4 + T cells. These molecules have different abilities to present viral antigens to CD4 + T cells. It has been proposed that diversity in HLA class II alleles may be involved in susceptibility or resistance to HCV infection (4, 5).
Many studies have shown that HLA class II gene polymorphisms may influence HCV infection. Studies of HLA gene polymorphisms and their associations with HCV outcome among various ethnic populations have had conflicting results. For example, the DRB1*0301 allele was associated with persistent HCV infection among German and Thai patients, whereas it was associated with HCV clearance in European and Korean people. The DQB1*0201 allele was associated with HCV clearance in Korean patients, while it was associated with persistent HCV infection in Thai patients (6). Therefore, we performed the present meta-analysis to derive a more precise estimation of the relationship between HLA class II alleles (HLA- DQ and HLA-DR) and the outcome of HCV infection.
2. Evidence Acquisition
2.1. Search Strategy
We searched databases (PubMed and Scopus) without timeline limits for English-language articles regarding HLA class II gene polymorphisms and their associations with HCV outcome. Our last search was conducted on May 15, 2021. In the present study for including related studies, we used various combinations of the following keywords: (1) “human leukocyte antigen”, (2) HLA, (3) “hepatitis C virus”, (4) “hepatitis C”, (5) HCV, and (6) spontaneous clearance.
2.2. Inclusion and Exclusion Criteria
Reports were regarded as qualified for inclusion if they met the following criteria: Reports with the full text available in the English language, reports with a proper study design such as case-control and cohort studies, reports that provided clear data about HLA class II gene polymorphisms, and their associations with the outcome of HCV infection, reports using high-resolution molecular typing (four-digit HLA typing ) method for the HLA class II genes, and studies that reported the frequencies of HLA-DRB1, HLA-DQA1, and HLA-DQB1 alleles in HCV-persistent infection and spontaneous clearance groups. Studies were excluded if they failed to present the data and results clearly and used low-resolution molecular typing (two-digit level) for HLA class II alleles. Furthermore, review articles, case reports, and case series were excluded from the assessment.
2.3. Data Extraction
In the current meta-analysis, two researchers (FS and AG) evaluated the quality of papers to select eligible studies. Following data were extracted from the included studies: The first author's name, year of publication, sample size, frequency of HLA class II alleles in spontaneous HCV clearance groups, and HCV-persistent infection groups. The analysis was performed as per the preferred reporting items for systematic reviews and meta-analysis.
2.4. Quality Assessment
The quality of the included studies was assessed using the Newcastle-Ottawa Scale (NOS). The score range of NOS is from 0 to 9 (7). If the NOS score of a study is ≥ 6, it can be considered high quality (8, 9).
2.5. Statistical Analysis
In this meta-analysis, we pooled the outcome estimate using Peto's method with a 95% Confidence Interval (CI). To assess the heterogeneity of the included studies, we used the Q test and I-squared statistic. We also used funnel plots to assess the publication bias. Publication bias was not assessed if the number of studies was < 10. The statistical analyses were performed using Stata software, version 11, and IBM SPSS statistics, version 22.
3. Results
3.1. Search Results and Study Selection
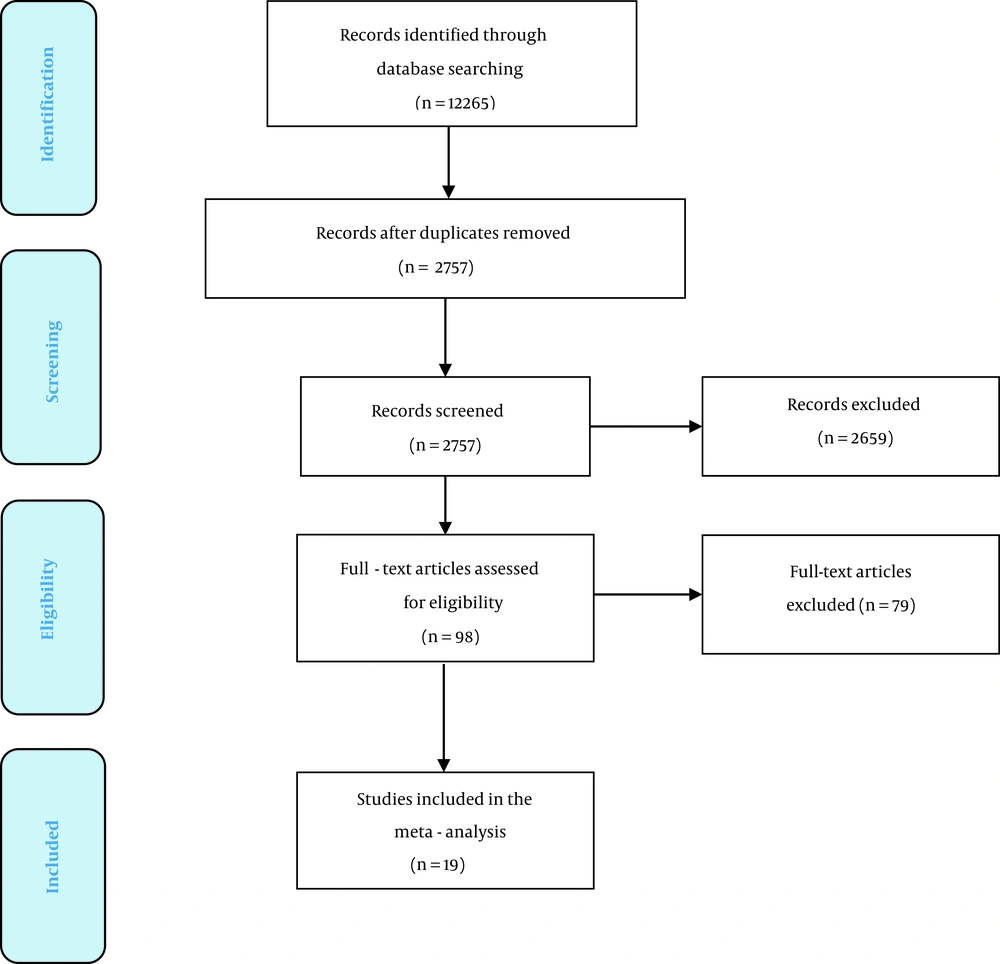
The paper selection process is illustrated in Figure 1. A total of 12,265 documents potentially related to the study objectives were identified through database searching, of which 2,757 papers were considered for the title and abstract screening after duplicate papers were excluded. Next, 2,659 documents were excluded based on title and abstract screening, and 98 articles remained. After the full-text screening, a total of 19 documents were found as eligible papers that evaluated the associations of HLA-class II genetic polymorphisms with HCV clearance. The included studies in this meta-analysis were published between 1997 and 2021. Table 1 lists the characteristics of the eligible and included studies. Regarding the effect of HLA-class II on the clearance of HCV infection, the studies largely evaluated the DQB1*0301 allele (17 studies), DQB1*0201 and DRB1*1101 alleles (14 studies), DQB1*0501, DRB1*0701, and DQB1*0302 alleles (13 studies), DQB1*0602, DQB1*0603, DRB1*0101, and DQB1*0502 alleles (12 studies).
| References | Year | Country (Ethnicity) | Spontaneous Clearance a (n) | Chronic HCV Infections b (n) | HLA Typing | HLA Class II Loci Studied | NOS |
|---|---|---|---|---|---|---|---|
| Alric et al. (10) | 1997 | France (European) | 25 | 103 | PCR-SSOP | DQB1, DRB1 | 7 |
| Cramp et al. (11) | 1998 | United Kingdom (European) | 49 | 55 | PCR-SSOP | DQA1, DQB1, DRB1 | 7 |
| Minton et al. (12) | 1998 | United Kingdom (European) | 35 | 138 | PCR-SSOP | DQB1, DRB1 | 7 |
| Lechmann et al. (13) | 1999 | Germany (European) | 9 | 18 | PCR‐SBT | DRB1 | 7 |
| Mangia et al. (14) | 1999 | Italy (European) | 35 | 149 | PCR-SSP | DQB1, DRB1 | 6 |
| Thursz et al. (15) | 1999 | Mix (European) | 85 | 170 | PCR-SSP | DQB1, DRB1 | 8 |
| Alric et al. (16) | 2000 | France (European) | 63 | 282 | PCR-SSOP | DQB1, DRB1 | 6 |
| Vejbaesya et al. (17) | 2000 | Thailand (Asian) | 43 | 57 | PCR-SSOP | DQA1, DQB1, DRB1 | 6 |
| McKiernan et al. (18) | 2000 | Ireland (European) | 95 | 148 | Reverse line probe hybridization | DQB1, DRB1 | 8 |
| Thio et al. (19) | 2001 | USA (American) | 200 | 374 | PCR-SSP | DQA1, DQB1, DRB1 | 6 |
| Azocar et al. (20) | 2003 | USA | 40 | 72 | PCR-SSOP | DQB1 | 6 |
| Spada et al. (21) | 2004 | Italy (European) | 10 | 24 | PCR-SSP | DQB1, DRB1 | 6 |
| Romero et al. (22) | 2008 | USA | 39 | 121 | PCR-SSP, PCR-SSOP | DQB1, DRB1 | 7 |
| Mangia et al. (23) | 2011 | Italy (European) | 49 | 68 | Reverse line probe hybridization | DQB1, DRB1 | 7 |
| Mangia et al. (24) | 2013 | Italy (European) | 47 | 122 | Reverse line probe hybridization | DQB1, DRB1 | 7 |
| Samimi-Rad et al. (25) | 2015 | Iran (Asian) | 54 | 63 | PCR-SSP | DQA1, DQB1, DRB1 | 7 |
| Huang et al. (26) | 2016 | China (Asian) | 231 | 429 | PCR‐SBT | DQB1, DRB1 | 7 |
| El-Bendary et al. (27) | 2019 | Egypt (African) | 108 | 235 | PCR‐SBT | DRB1 | 6 |
| Huang et al. (28) | 2019 | China (Asian) | 59 | 84 | PCR‐SBT | DQB1, DRB1 | 7 |
Characteristics of Studies Included in the Meta-analysis
3.2. Meta-analysis of Association of Class II HLA-DQ Locus Polymorphisms with HCV Clearance
The results of evaluating the class II HLA-DQ locus and its association with HCV clearance showed that subjects carrying HLA-DQB1*0301 and HLA-DQB1*0501 alleles were significantly associated with higher spontaneous HCV clearance (OR = 1.703, 95% CI: 1.464 - 1.981 and OR = 1.264, 95% CI: 1.021 - 1.565, respectively). Conversely, individuals carrying HLA-DQA1*0601, HLA- DQB1*0603, HLA-DQB1*0502, and HLA-DQB1*0201 alleles were associated with a lower probability of spontaneous HCV clearance (OR = 0.286, 95% CI: 0.083 - 0.986, OR = 0.627, 95% CI: 0.438 - 0.899, OR = 0.679, 95% CI: 0.495 - 0.933, and OR = 0.690, 95% CI: 0.578 - 0.824, respectively) (Table 2).
| HLA Alleles | Number of Studies | Spontaneous Clearance | Chronic HCV Infections | Odds Ratio (95% CI) | P Value | I2 Value (%) | Heterogeneity P Value | ||
|---|---|---|---|---|---|---|---|---|---|
| Number | Positive | Number | Positive | ||||||
| DQA1*0101 | 4 | 346 | 64 | 549 | 99 | 1.032 (0.729 - 1.461) | 0.859 | 45.6 | 0.138 |
| DQA1*0102 | 4 | 346 | 65 | 549 | 116 | 0.863 (0.615 - 1.212) | 0.442 | 0 | 0.566 |
| DQA1*0103 | 4 | 346 | 18 | 549 | 40 | 0.698 (0.394 - 1.239) | 0.265 | 0 | 0.703 |
| DQA1*0201 | 4 | 346 | 68 | 549 | 93 | 1.199 (0.848 - 1.696) | 0.326 | 74.5 | 0.008 |
| DQA1*0401 | 3 | 146 | 8 | 175 | 20 | 0.449 (0.192 - 1.053) | 0.074 | 0 | 0.681 |
| DQA1*0501 | 4 | 346 | 99 | 549 | 140 | 1.171 (0.866 - 1.584) | 0.314 | 52.9 | 0.095 |
| DQA1*0601 | 3 | 292 | 3 | 486 | 17 | 0.286 (0.083 - 0.986) | 0.036 | 55.4 | 0.106 |
| DQB1*0201 | 14 | 1052 | 219 | 2041 | 563 | 0.690 (0.578 - 0.824) | 0.000 | 52.7 | 0.011 |
| DQB1*0202 | 4 | 415 | 36 | 717 | 69 | 0.892 (0.585 - 1.361) | 0.671 | 71.1 | 0.016 |
| DQB1*0301 | 17 | 1164 | 416 | 2421 | 596 | 1.703 (1.464 - 1.981) | 0.000 | 57.5 | 0.002 |
| DQB1*0302 | 13 | 920 | 84 | 1829 | 167 | 1.000 (0.760 - 1.317) | 1.000 | 11.9 | 0.326 |
| DQB1*0303 | 10 | 786 | 67 | 1474 | 109 | 1.167 (0.850 - 1.603) | 0.365 | 0 | 0.897 |
| DQB1*0401 | 5 | 245 | 10 | 331 | 12 | 1.131 (0.481 - 2.662) | 0.828 | 0 | 0.726 |
| DQB1*0402 | 7 | 436 | 19 | 860 | 39 | 0.959 (0.547 - 1.681) | 1.000 | 0 | 0.917 |
| DQB1*0501 | 13 | 776 | 171 | 1538 | 281 | 1.264 (1.021 - 1.565) | 0.035 | 52.6 | 0.013 |
| DQB1*0502 | 12 | 885 | 57 | 1608 | 148 | 0.679 (0.495 - 0.933) | 0.018 | 0 | 0.887 |
| DQB1*0503 | 5 | 333 | 12 | 612 | 30 | 0.725 (0.366 - 1.436) | 0.411 | 5.4 | 0.376 |
| DQB1*0601 | 10 | 790 | 45 | 1365 | 72 | 1.085 (0.739 - 1.591) | 0.694 | 0 | 0.884 |
| DQB1*0602 | 12 | 953 | 112 | 1885 | 242 | 0.904 (0.712 - 1.148) | 0.434 | 31.2 | 0.142 |
| DQB1*0603 | 12 | 762 | 42 | 1528 | 130 | 0.627 (0.438 - 0.899) | 0.011 | 0 | 0.760 |
| DQB1*0604 | 10 | 624 | 29 | 1323 | 51 | 1.216 (0.763 - 1.937) | 0.463 | 0 | 0.824 |
| DQB1*0605 | 4 | 157 | 2 | 275 | 6 | 0.578 (0.115 - 2.901) | 0.716 | 0 | 0.983 |
| DRB1*0101 | 12 | 866 | 109 | 1667 | 132 | 1.674 (1.281 - 2.189) | 0.000 | 60.4 | 0.002 |
| DRB1*0102 | 5 | 342 | 10 | 664 | 19 | 1.023 (0.470 - 2.224) | 1.000 | 0 | 1.000 |
| DRB1*0103 | 2 | 144 | 8 | 203 | 12 | 0.936 (0.373 - 2.352) | 1.000 | 0 | 0.914 |
| DRB1*0301 | 11 | 995 | 129 | 1924 | 303 | 0.797 (0.638 - 0.995) | 0.048 | 41.8 | 0.063 |
| DRB1*0304 | 2 | 104 | 1 | 166 | 1 | 1.602 (0.099 - 25.892) | 1.000 | 0 | 0.356 |
| DRB1*0401 | 5 | 309 | 41 | 521 | 35 | 2.124 (1.321 - 3.416) | 0.003 | 56.2 | 0.058 |
| DRB1*0403 | 3 | 127 | 3 | 274 | 5 | 1.302 (0.306 - 5.533) | 0.712 | 0 | 0.616 |
| DRB1*0404 | 2 | 138 | 17 | 205 | 13 | 2.075 (0.973 - 4.424) | 0.078 | 0 | 0.802 |
| DRB1*0405 | 5 | 490 | 29 | 873 | 49 | 1.058 (0.659 - 1.698) | 0.809 | 59.7 | 0.042 |
| DRB1*0406 | 2 | 78 | 1 | 206 | 7 | 0.369 (0.045 - 3.051) | 0.453 | 0 | 0.714 |
| DRB1*0701 | 13 | 965 | 109 | 1929 | 276 | 0.763 (0.602 - 0.966) | 0.027 | 52.3 | 0.014 |
| DRB1*0801 | 2 | 104 | 1 | 118 | 3 | 0.372 (0.038 - 3.634) | 0.625 | 0 | 0.684 |
| DRB1*0803 | 2 | 274 | 21 | 486 | 33 | 1.139 (0.645 - 2.011) | 0.661 | 0 | 0.770 |
| DRB1*0901 | 7 | 744 | 52 | 1297 | 81 | 1.128 (0.787 - 1.618) | 0.515 | 0 | 0.997 |
| DRB1*1001 | 8 | 547 | 12 | 1019 | 22 | 1.016 (0.499 - 2.070) | 1.000 | 0 | 0.896 |
| DRB1*1101 | 14 | 1042 | 158 | 2150 | 201 | 1.733 (1.387 - 2.166) | 0.000 | 45.4 | 0.033 |
| DRB1*1102 | 2 | 209 | 7 | 392 | 10 | 1.324 (0.496 - 3.530) | 0.610 | 10.2 | 0.291 |
| DRB1*1103 | 2 | 235 | 1 | 523 | 5 | 0.443 (0.051 - 3.811) | 0.672 | 0 | 0.946 |
| DRB1*1104 | 6 | 384 | 30 | 794 | 63 | 0.983 (0.625 - 1.547) | 1.000 | 56.5 | 0.042 |
| DRB1*1201 | 8 | 430 | 29 | 965 | 34 | 1.980 (1.190 - 3.295) | 0.011 | 27 | 0.213 |
| DRB1*1202 | 2 | 274 | 29 | 486 | 51 | 1.010 (0.623 - 1.635) | 1.000 | 0 | 0.429 |
| DRB1*1301 | 10 | 560 | 32 | 1162 | 142 | 0.435 (0.292 - 0.648) | 0.000 | 24.9 | 0.214 |
| DRB1*1302 | 10 | 560 | 35 | 1162 | 122 | 0.568 (0.385 - 0.840) | 0.004 | 16.5 | 0.291 |
| DRB1*1303 | 5 | 245 | 8 | 630 | 7 | 3.004 (1.078 - 8.376) | 0.040 | 0 | 0.971 |
| DRB1*1305 | 2 | 52 | 4 | 75 | 6 | 0.958 (0.257 - 3.579) | 1.000 | 66.7 | 0.083 |
| DRB1*1401 | 7 | 412 | 15 | 966 | 47 | 0.739 (0.408 - 1.337) | 0.394 | 0 | 0.712 |
| DRB1*1501 | 8 | 582 | 65 | 1117 | 154 | 0.786 (0.577 - 1.071) | 0.147 | 0 | 0.595 |
| DRB1*1502 | 6 | 494 | 7 | 967 | 17 | 0.803 (0.331 - 1.950) | 0.828 | 0 | 0.531 |
| DRB1*1601 | 7 | 397 | 17 | 855 | 22 | 1.694 (0.889 - 3.227) | 0.116 | 0 | 0.837 |
| DRB1*1602 | 3 | 111 | 25 | 159 | 36 | 0.993 (0.556 - 1.774) | 1.000 | 0 | 0.404 |
Meta-analysis of Association of Class II HLA Alleles With HCV Spontaneous Clearance
3.3. Meta-analysis of Association of Class II HLA-DR Locus Polymorphisms with HCV Clearance
Among the studies analyzing the association of class II HLA-DR locus with HCV clearance, the meta-analysis identified that HLA-DRB1*1303 (OR = 3.004, 95% CI: 1.078 - 8.376), HLA-DRB1*0401 (OR = 2.124, 95% CI: 1.321 - 3.416), HLA-DRB1*1201 (OR = 1.980, 95% CI: 1.190 - 3.295), HLA- DRB1*1101 (OR = 1.733, 95% CI: 1.387 - 2.166), and HLA-DRB1*0101 alleles (OR = 1.674, 95% CI: 1.281 - 2.189) were significantly associated with higher spontaneous HCV clearance, whereas HLA-DRB1*1301 (OR = 0.435, 95% CI: 0.292 - 0.648), HLA-DRB1*1302 (OR = 0.568, 95% CI: 0.385 - 0.840), HLA-DRB1*0701 (OR = 0.763, 95% CI: 0.602 - 0.966), and HLA-DRB1*0301 alleles (OR = 0.797, 95% CI: 0.638 - 0.995) were associated with lower probability of spontaneous HCV clearance (Table 2).
3.4. Publication Bias Evaluation
In this study, the funnel plot was applied to assess the publication bias, and we did not find any evidence of publication bias (data shown in Supplementary File). However, the publication bias was not assessed for each HLA class II polymorphism with less than 10 studies.
4. Conclusions
The HCV infection is considered a multifactorial disease influenced by environmental and host factors. Host factors such as the innate and adaptive immune response play a crucial role in the outcomes of HCV infection (6, 29). It has been proposed that HLA molecules are the host factors that may influence the efficiency of the antiviral immune response to HCV infection. Various studies have confirmed that polymorphisms in HLA loci are associated with the outcome of HCV infection (6). Identifying these polymorphisms may help better understand the immune mechanisms of HCV clearance or persistence. The objective of this meta-analysis was to establish the updated information about the association of HLA class II alleles with clearance or persistence of HCV infection. The results showed that subjects carrying HLA-DQB1*0301, HLA-DQB1*0501, HLA-DRB1*1303, HLA-DRB1*1201, HLA-DRB1*0401, HLA-DRB1*0101, and HLA- DRB1*1101 alleles were significantly associated with higher spontaneous HCV clearance. These findings are almost similar to a previous systematic review conducted by Gauthiez et al. that reported HLA-DRB1*1101, HLA-DRB1*1201, HLA-DRB1*0101, and HLA-DQB1*0301 alleles were associated with HCV clearance (30). For HLA-DQB1*0501, HLA-DQB1*0603, and HLA-DRB1*1301 alleles, in contrast to our study, Gauthiez et al. reported a lack of association with HCV clearance (30). These inconsistent results may be due to the differences in the number of studies and the number of populations included in the meta-analysis. For example, for the HLA-DRB1*1301 polymorphism, we included four additional investigations in our meta-analysis compared to the meta-analysis published by Gauthiez et al. in 2017 (30). In addition, in 2005, a meta-analysis evaluated the association of HLA-DQB1*0301 and HLA-DRB1*1101 alleles with the outcome of HCV infection and reported that subjects carrying HLA-DQB1*0301 and HLA-DRB1*1101 alleles showed a reduced risk of developing chronic HCV infection (31), which is similar to this study and Gauthiez et al.'s meta-analysis (30). The results of our study also are in agreement with the results of studies by McKiernan et al. (18) and Thio et al. (19) that found the HLA-DQB1*0501 allele was associated with HCV clearance; however, in contrast to our study, Romero et al. reported that this allele was associated with persistent HCV infection (22). These contradictory results may be due to sample size and ethnic differences. It has been proposed that both viral genotype and host ethnicity can influence immunity against HCV infection. The relationship between HCV clearance and certain HLA alleles appears to be racially and geographically specific (19, 32). Therefore, well-designed studies in multiple regions are needed to confirm these results.
Some limitations exist in the present study that should be noted. First, the sample size in some of the included studies was small; therefore, further studies are needed to confirm the results. Second, the studies included in the present meta-analysis varied in the HLA typing technique, which may have affected the obtained results of this study. Third, some data were not included in the analysis since their original language was not English. Finally, some HLA class II polymorphisms were assessed in only two or three studies; therefore, more studies are needed to determine the association of HLA class II alleles with HCV outcome.
Taken together, the present meta-analysis provides detailed data on the association of HLA class II polymorphisms with HCV outcome. In the current meta-analysis, HLA-DQB1*0301, HLA-DQB1*0501, HLA- DRB1*1303, HLA-DRB1*1201, HLA-DRB1*0401, HLA-DRB1*1101, and HLA-DRB1*0101 alleles were significantly associated with higher spontaneous HCV clearance, whereas HLA-DQA1*0601, HLA-DQB1*0603, HLA-DQB1*0502, HLA-DQB1*0201, HLA-DRB1*1301, HLA-DRB1*1302, HLA-DRB1*0701, and HLA-DRB1*0301 alleles were associated with lower probability of spontaneous HCV clearance. Identifying an association between HLA class II polymorphisms and HCV outcome may open new insights to better understand the immune mechanisms of HCV clearance or persistence.