1. Background
Clinical medicine is now enjoying opportunities, which are brought by recent major progress in basic sciences. These advances might lead to improvement of health care by adapting a unique medical intervention at the right time for the right person. This approach, known as “personalized medicine”, is achievable through different levels, beginning from predicting the risk among healthy individuals to predicting the therapeutic response. Genomic information could provide a crucial base for guiding personalized medicine.
One of the best examples for this, though still remains controversial, is the result of genome-wide association studies showing that genetic polymorphisms near the interleukin 28B gene (IL28B; including rs12979860, rs12980275, and rs8099917) in patients with chronic hepatitis C, are associated with higher rates of sustained virological response to peginterferon alpha and ribavirin (1-5). Moreover, a relationship has also been reported between IL28B polymorphism and spontaneous clearance of HCV infection (1, 6, 7). The interferon, which is coded by IL28B ( i.e. IFN-λ) is known to play a role in defense against several viral infections, including hepatitis C and Hepatitis B Virus (HBV) (8). Therefore, the effect of IL28B polymorphism might not be specific to the immune response against infection with hepatitis C virus.
Results of studies evaluating the association between polymorphism of ILB28 and natural course of HBV or the response to treatment are divergent. Following negative results for a role for IL28B polymorphisms in spontaneous recovery of HBV (9, 10), large cohorts of patients with Chronic Hepatitis B (CHB) demonstrated that IL28B polymorphisms were independently related to serological responses to treatment with PEG-IFN (11, 12).
However, negative results on the spontaneous clearance of HBV were replicated (13) and studies indicated that the Single-Nucleotide Polymorphism (SNP) that has the strongest genetic association with HCV recovery (rs12979860) had no association with spontaneous recovery from HBV infection. Moreover, another study indicated an inverse influence of this SNP on HBV recovery (14). Thus, this polymorphism might have had a dissimilar effect on treatment outcome and spontaneous clearance of HBV infection. However, these results are not yet conclusive.
Despite the decline in HBV infection prevalence, the increasing overall number of chronic HBV infection is still a major health problem worldwide, including Iran (15-18). The situation calls for research in different populations to find the targeted treatment approaches. In the current study, the researchers investigated the influence of the IL28B polymorphism on HBV infection in a sample from the Iranian population.
2. Methods
2.1. Patients
The sample for this case-control study included Iranian Azeri patients with chronic hepatitis B virus infection (CHB) and those who Spontaneously Recovered (SR) from HBV infection. The protocol was confirmed by the regional ethnics committee, Tabriz University of Medical Sciences. All of participants provided written informed consent for participation and use of their genetic material for this study.
Between June 2015 and June 2016, patients with CHB infection, who visited the associated University clinic were recruited as the CHB group (HBsAg (+) for > 6 months, anti-HBc (+), and anti-HBs(-)). The SR subjects were selected from family members of HBV infected patients or those registered with the AZAR Cohort study (method described elsewhere) and were HBsAg (-), anti-HBc (+), and anti-HBs (+). A patient educational session was provided for all of participants.
All subjects were native Azeri Iranians. The exclusion criteria were as follows, (1) vaccinated subjects, who were positive for anti-HBs alone, but negative for anti-HBc, (3) co-infection with hepatitis C or human immunodeficiency viruses, and (4) presence of other chronic liver disease, such as primary biliary cirrhosis, autoimmune hepatitis, and decompensated liver disease. The researchers excluded 12 patients with chronic hepatitis B and 10 spontaneously recovered subjects. Fifteen patients from the spontaneously recovered group did not show, thus this study included 83 patients in the CHB group and 94 in the SR group.
2.2. Laboratory Tests
The HBV serological markers for HBsAg, anti-HBc, and anti-HBs were conducted with the Enzyme Linked Immunosorbent Assays (ELISAs).
2.3. Interleukin 28B Genotyping
Genomic DNA was extracted using a Ferments DNA Blood Kit (Ferments, France), according to the manufacturer’s instructions from peripheral blood samples. Quality control and subsequent handling of the DNA was carried out in the researchers own laboratory.
The genotyping of SNPs rs12979860 and rs8099917 was carried out by Polymerase Chain Reaction And Restriction Fragment Length Polymorphism (PCR-RFLP). For rs12979860, the primer sequences were 5’- GCG GAA GGA GCA GTT GCG CT -3’ (Rs12-F) and 5’- GGG GCT TTG CTG GGG GAG TG -3’ (Rs-12-R). For rs8099917, the oligonucleotide primers were 5’- CCC ACT TCT GGA ACA AAT CGT CCC -3’ (Rs80-F) and 5’- TCT CCT CCC CAA GTC AGG CAA CC -3’ (Rs80-F) (19).
For the RFLP assay, the PCR amplicon containing the rs12979860 SNP was digested with 10 units of Bsh1236I (BstUI) restriction endonuclease and the PCR amplicon containing the rs8099917 SNP was digested with 10 units of BseMI (BsrDI) restriction endonuclease for at least 1 hour.
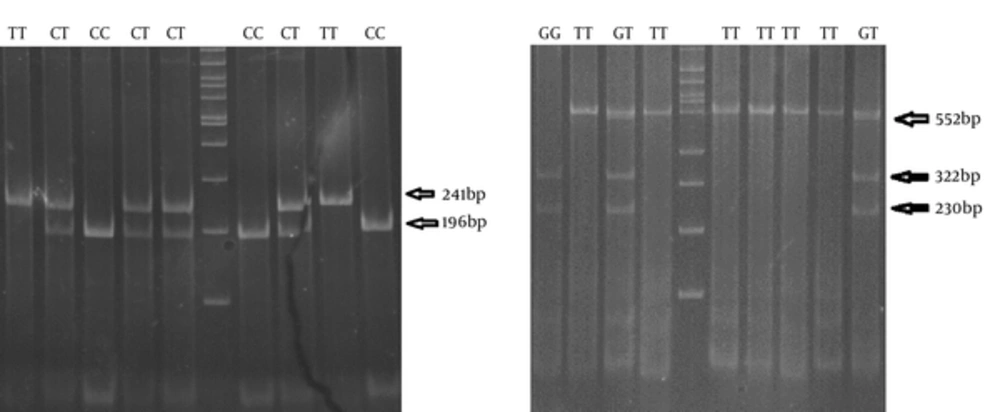
The digested PCR products were separated on 3% agarose gel, containing ethidium bromide and a visualized gel documentation system (Figure 1). The rs12979860 CC genotype produced two 196-bp and 45-bp fragments, the CT genotype produced three 241-, 196-, and 45-bp fragments and the TT genotype produced a 241-bp fragment. The rs8099917 TT genotype produced a 552-bp fragment, the GT genotype produced three 552-, 322- and 230-bp fragments and the GG genotype produced two 322- and 230-bp fragments.
2.4. Statistical Analysis
Data are expressed as the means (standard deviation) and medians (range) for age, and numbers (%) for genotype. The laboratory findings were compared among the groups using Chi-square tests and SPSS version 17.0 (SPSS, Inc., Chicago, IL). A P value of < 0.05 on a two-tailed test was considered statistically significant.
3. Results
The median age of the subjects in the SR (52 males and 42 females) and the CHB groups (47 males and 36 females) was 38 and 44 years, respectively. The mean and Standard Deviation (SD) was 37.5±10.3 and 39.1±9.8, respectively. Gender distribution was not different between the 2 groups (P = 0.812).
3.1. Prevalence of rs12979860 and rs8099917
Regardless of group, rs12979860 CT was identified in more than 50% of participants (65.0% in the SR, and 52.5% in the CHB group). The rs8099917 TT was also the most prevalent (56.4% in the SR and 74.7% in the CHB group; 65% total prevalence).
Distribution of rs12979860 polymorphisms was not different between the 2 groups (P = 0.224). However, the prevalence of TT rs8099917 was significantly higher in the CHB group than in the SR subjects (P = 0.038, OR = 1.435 (1.1 to 1.872)). Results are shown in Table 1.
| Variables | Spontaneous Recovery | Chronic Hepatitis B |
|---|---|---|
| rs12979860 | ||
| No. | 94 | 83 |
| CC | 28 (29.8) | 31 (37.3) |
| CT | 49 (52.1) | 44 (53.0) |
| TT | 17 (18.1) | 8 (9.6) |
| C allele | 105 | 106 |
| T allele | 83 | 60 |
| rs8099917 | ||
| No. | 94 | 83 |
| GG | 9 (9.6) | 4 (4.8) |
| GT | 32 (34.0) | 17(20.7) |
| TT | 53 (56.4) | 62 (74.7) |
| G allele | 50 | 25 |
| T allele | 138 | 141 |
Prevalence of Different Genotypes in Selected Loci Near the IL28B Gene in the Study Sample
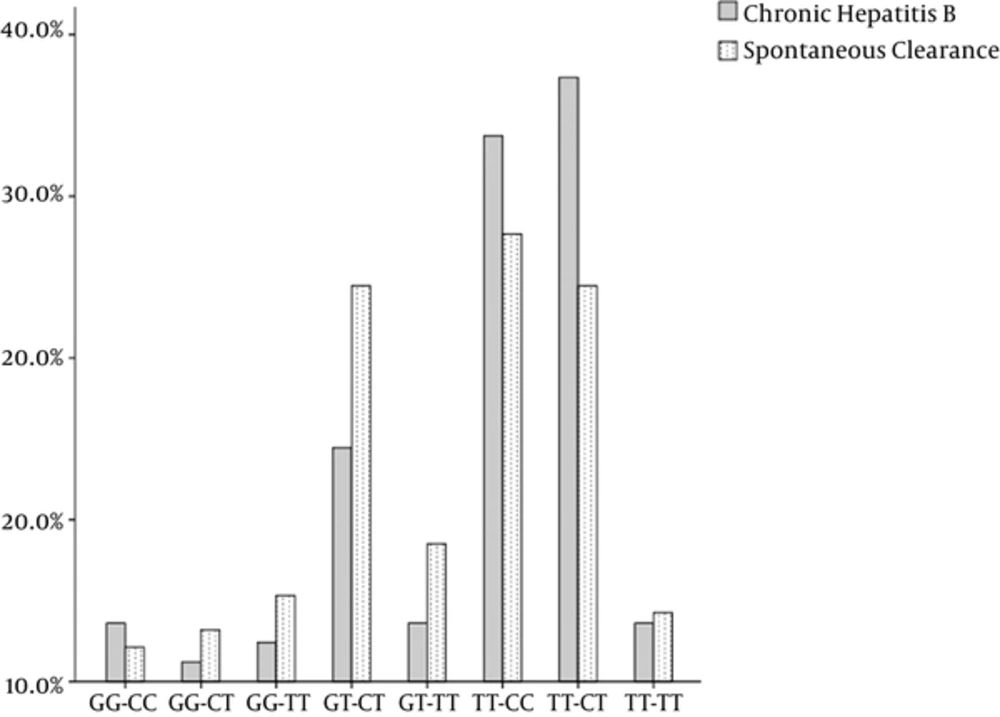
3.2. Combination of Genotypes
Frequencies for combination of genotypes using rs12979860 and rs8099917 are presented in Figure 2. As shown in this all all types of combinations were observed in both groups, except for rs8099917 GT- rs12979860 CC. This also also shows that rs8099917 TT- rs12979860 CT is the most prevalent in the CHB group and rs8099917 TT- rs12979860 CC is the most prevalent in the SR group.
Combination of genotypes did not show statistically better odd ratios (all P > 0.05).
4. Discussion
Results of this study indirectly showed the pattern of IL28B upstream polymorphisms in the Iranian population. In several studies from Hong Kong (20), Australia (21), Korea (13, 14), and China (22), rs12979860CC was the most common genotype regardless of hepatitis infection status with a noticeable difference to other genotypes. This difference decreased in studies from Arabian countries (23) where rs12979860CC was still foremost. On the other hand, rs12979860CT is the most common genotype reported by studies from Turkey (24, 25), similar to the current results from Iranian Azeri-Turk population. However, this similarity is not restricted to inhabitants of Azerbaijan province yet are reported from other parts of Iran as well (19, 26). Fewer data are available about polymorphism in rs8099917, and comparable to the current results; rs8099917TT is the most common (13, 14, 20). Though this dominance is more obvious in some report, rs8099917GG was not detected in a Korean sample (27).
In the current study, the researchers genotyped IL28B polymorphisms (rs8099917 and rs12979860) in Iranian Azeri patients with chronic hepatitis B and those who spontaneously recovered from HBV infection, to determine its possible influence on HBV infection outcome in a natural history setting. The current results are in line with previous studies indicating a different role for IL28B polymorphisms in the natural course of HBV infection compared to HCV.
In contrast to a previous study (9), IL28B polymorphisms (rs8099917) was significantly associated with the outcome of HBV infection in the current sample. This result has previously been reported by other studies as well (14). Conversely, the prevalence of the favorable predictor in terms of spontaneous recovery from HCV infection (i.e. rs12979860CC) was not related to natural course of HBV in the current sample. This finding has also been reported in several previous studies from other populations (28).
Product of IL28B gene, is a cytokine that ultimately up-regulates the IFN-stimulated genes (29, 30). Though the mechanism is not clear, this cytokine plays a critical role in the immune response to HCV infection and increasing evidences show its effect on outcome of both natural and IFN-α-treated HCV infections (1, 31). However, there is still controversy on the topic of the association between IL28B genetic polymorphism and both natural and treated HBV infection (32).
Following the first positive report about the association between IL28B polymorphism and chronic HBV infection (12), which proposed that rs12980275AA is independently associated with HBeAg seroconversion after PEG-IFN treatment in CHB, there have been several negative results.
One study concluded that the rs12979860CC genotype, which has the strongest genetic association with HCV recovery, was not associated with spontaneous recovery of HBV infection (9). Another study reported a significant but reverse association compared to HCV results and showed higher prevalence of rs12979860CC and rs8099917TT in patients with CHB compared to spontaneously recovered subjects (14). This is why in patients with HCV infection, rs12979860CC is associated with a 2- to 3-fold higher response to treatment (1) and spontaneous clearance (6), and there are evidences that presence of rs8099917TT might increase this association (33). As a result, the mechanism of immune response in which product of this gene takes a part, might be different for HBV and HCV infections. This may be a result of differences between HBV and HCV infection (34), characteristics of IL28B (35) or a combination of both. However, this hypothesis and the current results should be replicated in future studies. The present result could also be included in prospective studies so that together with other biomarkers that might effect the course of illness (36), they could end with a precised method of intervention for each patient.
In conclusion, the SNP upstream of IL28B has an important role in natural course of HBV infection that might be different from HCV infection.