1. Background
Branched-chain aminotransferase 1 (BCAT1) is a major enzyme initiating the degradation of branched-chain amino acids (BCAAs), including leucine, isoleucine, and valine, to form the corresponding branched-chain alpha ketoacid and amino acids, and provide energy and raw materials for other metabolic pathways in the body (1). BCAT1 is only expressed in a few tissues, such as the brain, and is not expressed in the liver. In addition, compared to the matching normal tissues, it is highly expressed in various human malignancies (2). The expression of BCAT1 in the liver cancer tissues was related to tumor differentiation. BCAT1 is abundant in poorly differentiated hepatomas. However, the expression of BCAT1 on a variety of benign liver cancer cells was close to normal tissues, such as hepatoma cells 7794A, 7316A, and 7777, which did not express BCAT1. On the other hand, the highly malignant hepatoma cell lines, including 5123tc and 7793, expressed high levels of BCAT1 (3, 4). Moreover, it has been documented as a key prognostic predictor in HCC. Our previous investigation showed that BCAT1 enhances HCC cell growth in vitro and in vivo, and the expression of BCAT1 is remarkably linked with tumor differentiation (5, 6). Gabapentin, originally an antiepileptic medicine, is more commonly used to treat neuropathic pain. Recently, it has been found to have significant benefits in non-neuropathic pain, especially perioperative and cancer pain (7). Further studies have shown that gabapentin can inhibit BCAT1 in the cytoplasm (8). Tonjes et al. reported the anti-proliferation influence of gabapentin in glioma cell lines, and Hattori et al. found that blocking BCAT1 gene expression can induce cellular differentiation and dampens the propagation of chronic myeloid leukemia (CML) in vitro and in vivo (9, 10). Gabapentin, as a chemical inhibitor of BCAT1, can effectively inhibit the clonal growth of human CML cell lines (10). This suggests that gabapentin may potentially inhibit tumor propagation by inhibiting BCAT1.
2. Objectives
Herein, we assessed the influence of gabapentin on the growth, migration, and tumorigenesis of hepatoma cells. Moreover, the underlying molecular mechanisms of gabapentin on HCC cells were preliminarily evaluated by RNA-seq.
3. Methods
3.1. Reagents
1-(Aminomethyl)-cyclohexane acetic acid, Neurontin (Gabapentin) was provided by Sigma-Aldrich (St. Louis, United States). The storage concentration of gabapentin was 500 mM and was dissolved in PBS before use. RPMI 1640 medium, anti-microbials, and trypsin-EDTA were supplied by Gibco (Thermo Fisher Scientific, Waltham, United States).
3.2. Cell Culture
HCC cell lines Bel7404, Hep3B, Huh7, and SMMC-7721 were inoculated in RPMI 1640 enriched with 10% FBS in 5% CO2 and 37°C.
3.3. Cell Proliferation Assay
The proliferation of cells was assessed by the MTT assay (Cell Proliferation Kit I, Roche, Germany) as described by the manufacturer. An enzyme-linked immunosorbent assay reader was utilized to read the ODs at 650 nm and 595 nm as the reference and test wavelengths, respectively.
3.4. Transwell Cell Migration Assay
Migration assay wells of 6 mm diameter with a membrane pore size of 8 µm were used in this study. From the 25 cm2 stock flasks, cells were digested by 0.25% EDTA-free pancreatin and were adjusted to a concentration of 5 × 108 cells/mL. A volume of 100 µL of the cell suspension was aspirated and placed into the top compartment of each well, and with a 600 µL complete culture medium containing 10 mM gabapentin was added into the lower chamber. A complete culture medium without gabapentin was introduced to the lower control wells and then allowed to grow for 12 h at 37°C. Thereafter, the membrane inserts were removed and fixed with 4% polyoxymethylene. The number of cells that migrated in the presence or absence of gabapentin is shown by cells on the underside of the membrane. After allowing the insert to dry, crystal violet was added to the cells, and each membrane was photographed under an inverted fluorescence microscope (Olympus, Tokyo, Japan) for analysis. Five random microscope fields (200×) were counted for each chamber, and two independent experiments were performed.
3.5. RNA-seq and Analyses
RNA sequencing was performed by ShBio Inc. (Shanghai, China). Total RNA was isolated with the RNAiso Plus Total RNA extraction reagent (TAKARA; Cat#9109) as described by the manufacturer. We checked for the integrity of the isolated RNA on the Agilent 2100 Bioanalyzer Platform (Agilent Technologies, Santa Clara, United States). Afterwards, RNA purification was performed with the RNAClean XP Kit (Beckman Coulter, Inc. Kraemer Boulevard Brea, CA, USA; Cat A63987) and RNase-Free DNase Set (QIAGEN, GmbH, Germany; Cat#79254). RNA sequencing was completed by processing the mRNA library in which the mRNA samples were fragmented and the generation of first-strand cDNA. Next, we generated second-strand cDNA, followed by the end repair of cDNA. Afterwards, we adenylated the 3’ends a single, followed by the ligation of adapters. We enriched the ds-cDNA fragments and validated the enriched library. The cDNA libraries were measured by Qubit® 2.0 Fluorometer and QubitTM dsDNA HS. The size and purity of the library were detected by the FlashGelTM Dock system combined with FlashGelTM DNA Cassette. The peak value of the library is generally about 350 bp. Formation of clusters, hybridization of primers, and sequencing were completed as described by the manufacturer. The second procedure of RNA sequencing was conducted on the Illumina® Hiseq2500. The expression levels for each gene were then normalized to fragments per kilobase of transcript per million mapped reads (FPKM). FPKM=total exon fragments/mapped reads in millions×exon length in kb. Differential expression analysis of each gene between two groups was performed using edgeR. The false discovery rate (FDR) of each gene was assessed based on the mean log2 (fold change [FPKM of gabapentin/FPKM of control]). Genes remarkably expressed differently between two samples which were identified with a P-value cutoff of 0.05. As a criterion for differentially expressed genes (DEGs) constituted a fold change greater than 2.
GeneSpring was utilized to create DEG clustering via Pearson correlation and the average linkage approach. Database for Annotation, Visualization, and Integrated Discovery (DAVID) was utilized to assess the functional clustering of DEGs. Hypergeometric tests and correction using the Benjamini-Hochberg procedure were used to determine the remarkable GO terms coupled with the KEGG pathway of DEGs. P-value < 0.01 along with enrichment gene count of > 2 were applied to assess the remarkable GO biological process, while P < 0.05 with enrichment gene count of > 2 was adopted to examine the enriched KEGG pathways.
3.6. Mouse Xenograft Tumor Models
All mouse procedures were performed in compliance with the National Institute of Standards and Practices for the Care and Use of Laboratory Animals. The University Committee on the Ethics of Animal Experiments authorized the animal protocols. Six-week-old BALB/c nude mice were acquired from Shanghai Laboratory Animal Center (SLAC) and were maintained in a dedicated pathogen-free setting for the in vivo tumor growth experiment. For subcutaneous xenograft model experiments, Huh7 cells (2.5 × 106) were subcutaneously inoculated into the back of each mouse with a hypodermic needle. The mice were divided randomly into the vehicle-control and treatment study groups (n = 6/group). Following randomization, the mice in the experimental group received 0.5 mL of gabapentin 20 mM intraperitoneal (IP) inoculation every other day for a week, making three IP injections. The control group had the same schedule but was inoculated with an equivalent volume of saline. The size of the tumors was assessed once a week with precision calipers. The volume of the tumor mass (xenograft) was determined using the following formula: volume=[(tumor length)×(tumor width)2]/2. On day 21 of randomization, the mice were sacrificed, and the tumors were removed, weighed, and photographed.
3.7. Quantitative Real-time PCR
Total RNA was isolated with TRIzol Reagent (Invitrogen, Carlsbad, United States). Thereafter, cDNA was generated with RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) and the Taq PCR Master Mix (Takara, Dalian, China). Afterwards, qPCR was conducted with SYBR® Green real-time PCR Master Mix on the ABIPrism® 7500 Sequence Detection Platform (Applied Biosystems, Foster City, United States). The primers used to amplify CDH11, ARHGAP15, and GAPDH transcripts are given in Table 1. The data were analyzed using the SDS software version 1.4.
| Gene | Forward | Reverse |
|---|---|---|
| GAPDH | GTCTTCACCACCATGGAGAA | TAAGCAGTTGGTGGTGCAG |
| CDH11 | CCACGGCTCCTCCTTATGAC | AATCTGTGGTGGCCGACTCT |
| ARHGAP15 | TCCATGATCCTCACCGATGT | TCAACCATCAGTTGTTCCAATG |
3.8. Statistical Analysis
All quantitative data are given as mean ± SD of triplicate measurements from one of two independent experiments, which gave similar results, and P-value < 0.05 was considered statistically significant. Continuous variables were compared by the student’s t-test. All data analyses were performed by the SPSS software version 16.
4. Results
4.1. Gabapentin Reduces the Proliferation of Human Hepatoma Cells
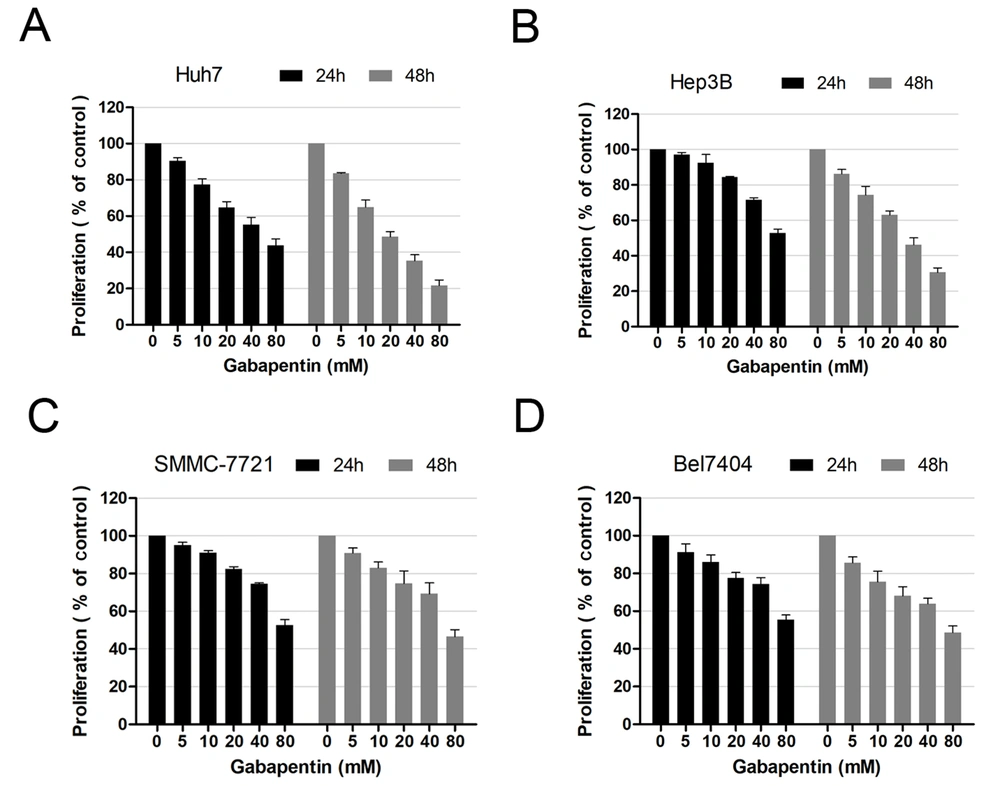
To examine the effects of gabapentin on hepatoma cells, we compared the impacts of gabapentin on four HCC cell lines (Bel7404, Hep3B, Huh7, and SMMC-7721) in vitro. The MTT assay at 24 and 48 h was performed to determine growth inhibition after exposure to different concentrations of gabapentin (0, 5, 10, 20, 40, and 80 mM). The cell survival rate of various cell lines decreased gradually with an increase in gabapentin concentration. In contrast to the controls, a remarkable difference was observed in the concentration of gabapentin beginning from 20 mM (Figure 1).
Moreover, it was observed that the cell survival rate at 24 h was higher than that of 48 h after treatment with the same gabapentin concentration. These data demonstrated that the inhibitory effect of gabapentin on human hepatoma cells was both dose- and time-dependent.
4.2. Gabapentin Inhibits the Migration Properties of Human Hepatoma Cells
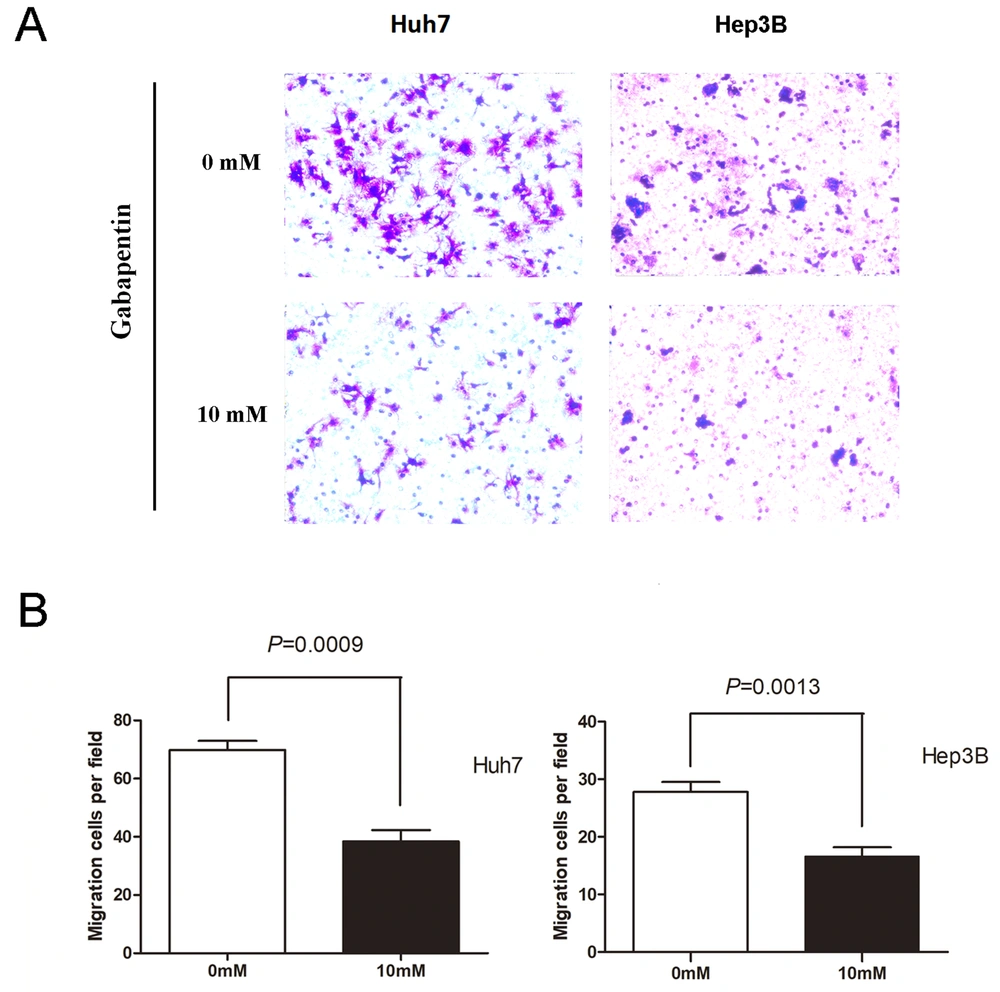
Logarithmic growth phase Huh7 and Hep3B cells were used as experimental cells, and FBS was used as chemokine throughout the transwell migration experiment. The cell complete culture solution containing 10 mM gabapentin in the lower chamber was set up for the experimental group. The cell complete culture solution without gabapentin in the lower room was regarded as the control group. Cell migration was examined as described in materials and methods. As illustrated in Figure 2, cell migration attracted by FBS was remarkably dampened in two different types of human hepatoma cell lines that were inoculated with 10 mM gabapentin. The Huh7 cell study group had a much lower number of cells migrated than the controls (69.80 ± 7.09 vs. 38.40 ± 8.79, P = 0.0009). Similar results were observed in the Hep3B cell experimental group (27.80 ± 3.83 vs. 16.60 ± 3.51, P = 0.0013). These data indicate that gabapentin can inhibit the migration of hepatoma cells.
4.3. Gabapentin Inhibits the Tumorigenicity of Human Hepatoma Cells In Vivo in Nude Mouse Xenografts
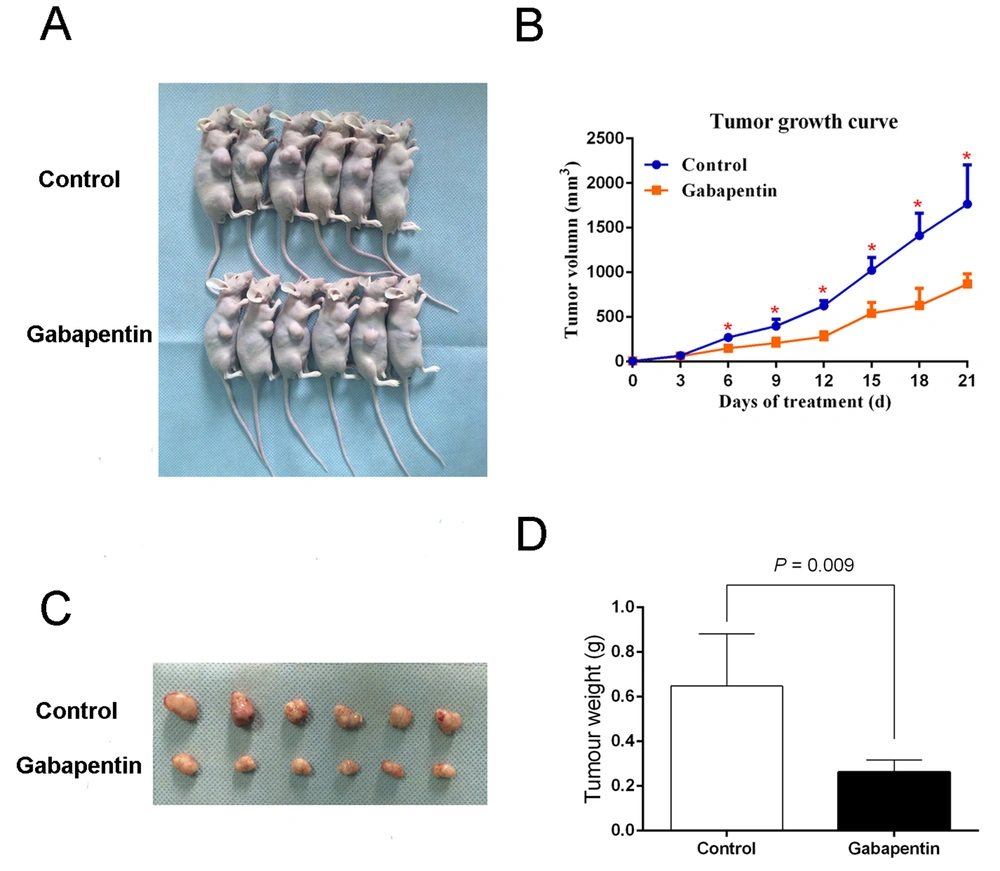
We inoculated nude mice subcutaneously with 2.5×106 Huh7 cells. Each nude mouse in the experimental group was injected IP with 0.5 mL gabapentin 20 mM every other day for 7 days. The control group animals were injected IP on alternate days with the same volume of saline. The mice were fed normally over 21 days from randomization, and no deaths occurred during this time. The mice were sacrificed at the end of this period, and the tumor was harvested. The mean weight of tumor per mouse in the control group was 0.65 ± 0.23 g, and the mean tumor volume was 866.33 ± 281 mm3. The mean weight and tumor volume in the experimental group were 0.26 ± 0.05 g and 408.24 ± 84.09 mm3, respectively. The tumor weight along with the volume of the experimental group were both remarkably lower than the controls, P = 0.009 (Figure 3), suggesting that gabapentin inhibits the tumorigenicity of HCC cells in nude mouse xenografts.
Tumorigenicity assay in nude mice with gabapentin treatment. (A) Nude mice with tumor formation. (B) Tumor growth curve in nude mice. Measurement of the size and volume of tumors every 3 days. (C) Images of tumors derived from Huh7 cells with gabapentin treatment and saline in nude mice. (D) Weight of tumors.
4.4. RNA-sequencing Profile in Huh7 Cells After Gabapentin Treatment
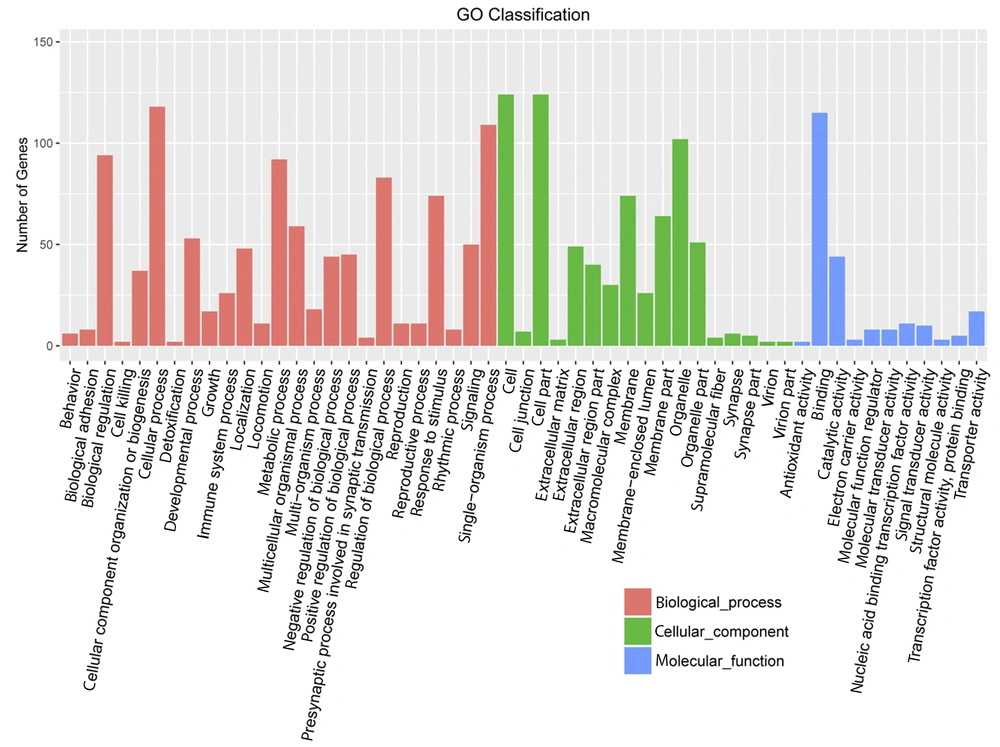
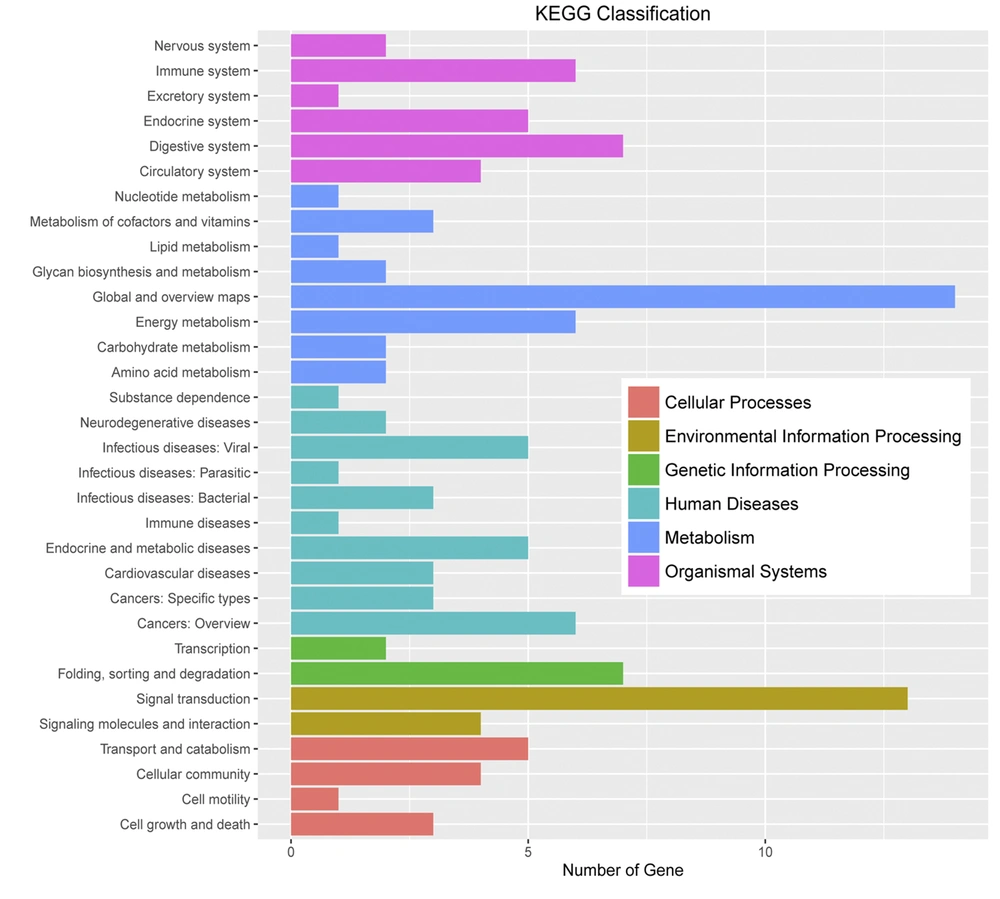
Huh7 cells were used as experimental cells. The Huh7 cells treated with gabapentin were used as experimental group, which treated with 20 mM gabapentin for 24 h. However, the normal Huh7 cells without gabapentin treatment were used as the control group. Based on the DEG selection criterion (i.e., FDR less than 0.05 and more than 2-fold change between experimental and control cells), DEGs were identified. Initial characterization of DEGs showed that they were mainly concentrated in “biological modulation”, “cell process”, “metabolic process”, “modulation of biological process”, “single-organism process”, “cell”, “cell part”, “organelle”, and “binding” in gene ontology terms and “immune system”, “digestive system”, “energy metabolism”, “cancers”, “folding, sorting, and degradation”, and “signal transduction” in KEGG pathway enrichment analysis (Figures 4 and 5).
4.5. Effects of Gabapentin on the Expression of Some Genes in Hepatoma Cells
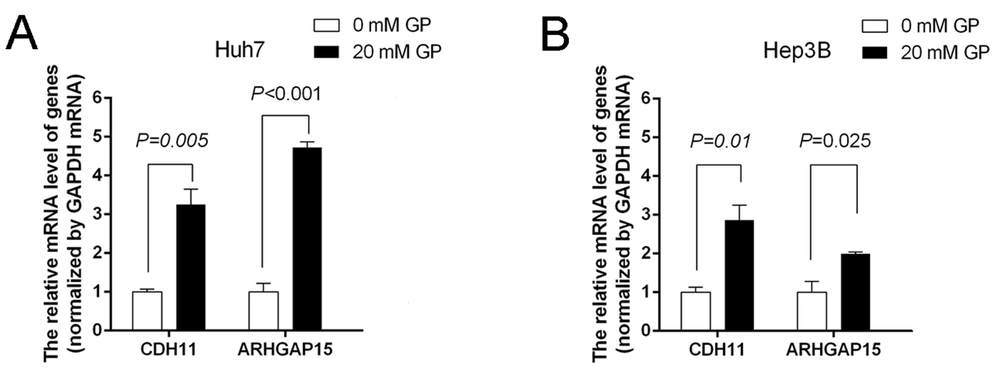
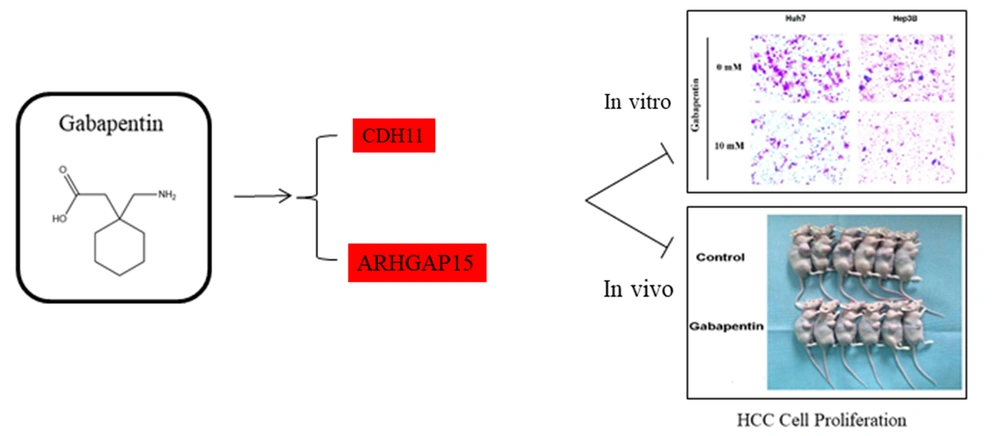
According to the results of RNA-seq data, we screened some genes associated with “signal transduction” and “cancers”, including CDH11 and ARHGAP15, for further qRT-PCR validation. Compared to the controls, the relative expression of CDH11 and ARHGAP15 in the gabapentin-treated group was significantly upregulated (P < 0.05) (Figure 6A and B). These results showed that the inhibitory effect of gabapentin on hepatoma cells might be associated with the signaling pathways of CDH11 and ARHGAP15 genes (Figures 7).
5. Discussion
The BCAAs are essential amino acids that affect the protein synthesis, which needed by the human body and tumor tissues (11). Due to the interaction between tumor and host, multiple types of tumors are accompanied by obvious changes in BCAAs metabolism (12, 13). Supplementation of BCAAs is critical in the clinical treatment of HCC. Studies found that after radio-frequency ablation, HCC patients with BCAAs supplementation had increased event survival time and reduced complications (14). Clinical trials also found that supplementation of BCAAs can improve the prognosis of patients with HCC (15, 16). BCAT1 has attracted attention as a key enzyme of BCAAs catabolism. A previous investigation indicated that the over-expression of BCAT1 correlated significantly with the serum levels of AFP and tumor differentiation in HCC patients. Furthermore, dampening BCAT1 expression promoted cisplatin-triggered cell death and BCAT1-triggered cisplatin sensitivity. BCAT1 dampening has been proposed as an available treatment approach for enhancing the chemotherapy effects and improving clinical outcomes in HCC subjects (5).
Gabapentin is an analog of γ-GABA, similar to GABA in structure. It was first approved for the clinical treatment of epilepsy, and subsequently, it has been widely used in easing pain, such as neuropathic pain, cancer-related pain, and perioperative pain (17). In addition to cancer pain, gabapentin can also alleviate other cancer-related symptoms, including improving the deglutition of oropharyngeal squamous cell carcinoma during the chemotherapy period and relieving hot flushes in patients with prostate cancer (18, 19). It was found that gabapentin can inhibit BCAT1 activity by binding to it in the cytoplasm. Furthermore, researchers have used gabapentin as a BCAT1 inhibitor to treat CML. Using gabapentin as a BCAT1 inhibitor to block the activity of the BCAT1 enzyme can induce cell differentiation in vitro and in vivo, reducing the reproduction of CML (10). These findings suggest the potential use of gabapentin as a BCAT1 inhibitor for cancer therapy. Among the anti-cancer reports, studies have shown that metformin and gabapentin salt has a good inhibitory effect on triple-negative breast cancer (20). There are no previous reports of using gabapentin in HCC. However, we recommend that, as a BCAT1 inhibitor, it might play an indispensable role in HCC treatment.
Herein, it was shown that gabapentin could dampen the proliferation of various hepatoma cells in a time- and dose-dependent approach. After treatment with 10 mM gabapentin for 12 h, the migration number of hepatoma cells through the transwell intima reduced significantly. The results of tumor graft growth showed that the mean tumor volume and weight in the gabapentin-treated group were remarkably lower than in the normal saline control group. Therefore, we believe that gabapentin had a dampening effect on the growth and proliferation of hepatoma cells. The previous study showed that gabapentin could diminish cell growth independent of BCAT1 (21). Nonetheless, evidence has documented gabapentin as a repressor of BCAT1, and gabapentin is still commonly used to inhibit the BCAT1 enzyme as an inhibitor in recent studies (22, 23). The mechanism through which gabapentin dampens HCC cell growth needs further exploration. The RNA-Seq results suggested that DGEs are mainly concentrated in “cancer”, “energy metabolism”, “signal transduction”, and other important biological pathways. Compared to the control group, some remarkable genes, including CDH11 and APHGAP15, were upregulated, which was previously reported to be related to the proliferation and invasion of cancer cells (24, 25). Furthermore, we verified these genes by qRT-PCR and showed that the relative expression of CDH11 and ARHGAP15 in hepatoma was significantly upregulated after treatment with gabapentin. Therefore, we suggest that the inhibitory effect of gabapentin on hepatoma cells may be linked to the signal pathways of these genes.
As an analgesic widely used in clinical trials, there is little evidence for the carcinogenic effect of gabapentin in oral/pharyngeal cancer and liver cancer through case-control analyses using conditional logistic regression and odds ratio estimation of relative risk using criteria from previous collaborations (26-28). Liver cancer is highly malignant, and sorafenib has been the only first-line treatment for advanced HCC since 2008. However, several clinical studies have shown that the overall efficacy of cytotoxic chemotherapy is far from satisfactory, leading us to consider whether to use other mechanisms to inhibit the growth of HCC cells (29-31).
In the present research, we conducted a preliminary exploration of the inhibitory effect of gabapentin on hepatoma cells and its mechanism. It has been observed that gabapentin has an inhibitory effect on the cell growth, motility, and tumor formation of hepatoma cells, which will lay a foundation for future studies on BCAT1 as a target of HCC treatment. Furthermore, this paper has some limitations due to the novelty and uncertainty of gabapentin in treating liver cancer. Basic research findings from these studies need to be assessed by clinical research and still need further investigation. The clinical use of gabapentin treating HCC as a target of BCAT1, its clinical therapeutic effect, and specific molecular mechanisms need further verification.