1. Background
Hepatitis B virus (HBV) infection is a major global health problem and the most severe form of viral hepatitis, leading to cirrhosis and liver cancer when chronic. According to the World Health Organization, a fact sheet published on 24 June 2022 showed that approximately 296 million individuals were living with chronic HBV infection in 2019, with 1.5 million new infections each year.
During the infection, antibody responses play an essential role in eliminating HBV particles and infected hepatocytes. Infected individuals can naturally clear hepatitis B surface antigen (HBsAg) and produce hepatitis B surface antibodies (anti-HBs); however, others might progress to cirrhosis and hepatocellular carcinoma (HCC) (1, 2).
The serological diagnosis of HBV infection is primarily based on testing for surface antigen (HBsAg), the major glycoprotein of the viral envelope. Various serological markers for HBV infection include HBsAg, anti-HBs, HBeAg, anti-HBe, and anti-HBc immunoglobulin M and Immunoglobulin G. Various serological markers allow for identifying individuals infected with HBV, studying the progression of chronic hepatitis B, and monitoring antiviral treatment (3). The presence of anti-HBs is considered an indicator of immunity to HBV infection. On the other hand, the absence of HBsAg is a criterion for excluding infections. Therefore, the HBsAg test is often used to screen blood and organ donors (4, 5).
It has been shown that the region between amino acids 124 and 147 of protein S represents the “a” determinant in the major hydrophilic region (MHR) of the gene encoding HBsAg (6), which is common to all variants of the HBsAg gene. In general, the mutation in the “a” determinant causes the induction of the generation of HBV immune escape mutants (7-9).
Although most studies on HBsAg escape mutants have specifically targeted the MHR, further research has shown that mutations outside the MHR can also give rise to escape mutants (10, 11). In addition, mutations near the MHR can alter the specific antigenicity of the group even if the mutations are not within the MHR (6, 12, 13).
In serological markers of HBV infection, general theory considers that the antibody to HBsAg (anti-HBs) can neutralize HBsAg; therefore, it is generally agreed that there will be no simultaneous positive for both HBsAg and anti-HBs in routine clinical practice (14-16). However, the simultaneous presence of HBsAg and anti-HBs in chronic HBV carriers has been reported in previous studies (17, 18). Although there are several reasons for the simultaneous presence of HBsAg and anti-HBs in the sera of patients with chronic hepatitis, the outcome of this coexistence for patients remains largely unknown. Moreover, viewpoints are still controversial, and the mechanism underlying this serological pattern needs to be clarified.
2. Objectives
This study examined patients with chronic HBV infection to determine the coexistence of HBsAg and anti-HBs and analyze the infection’s clinical and virological features. In addition, this study investigated mutations in the MHR of HBsAg.
3. Methods
3.1. Patients and Samples
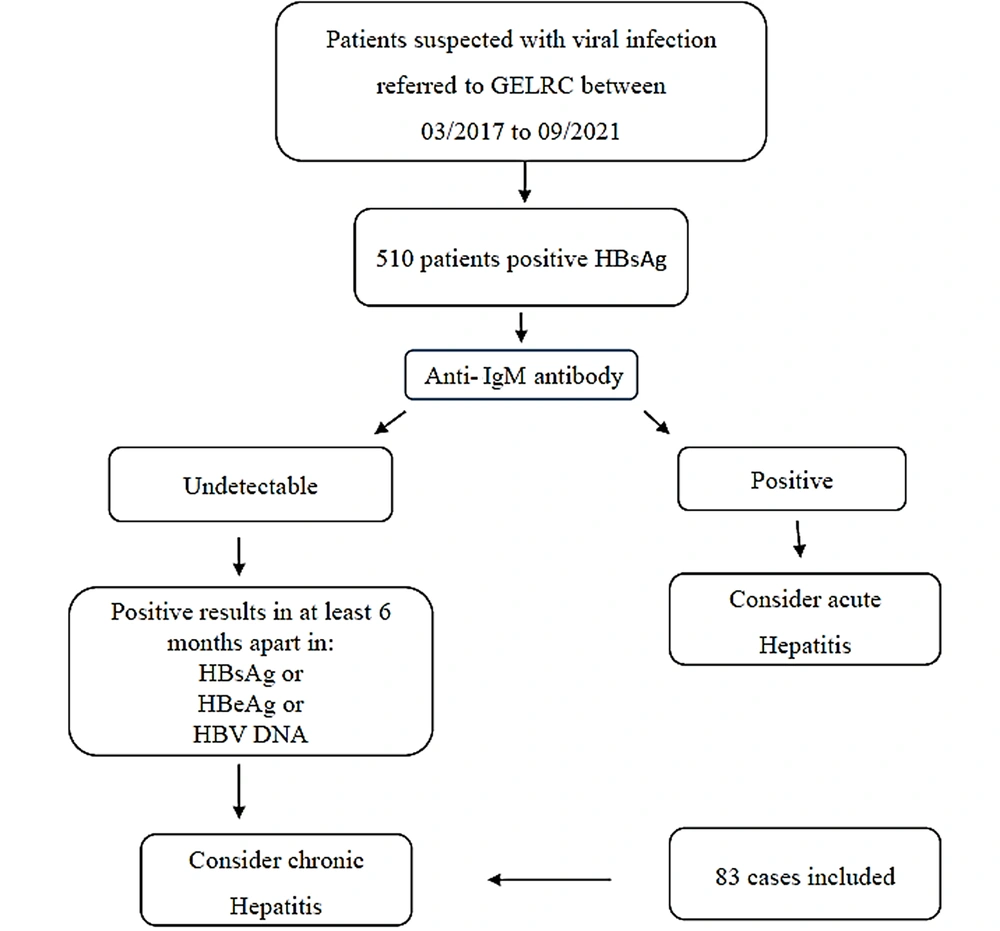
Within March 2017 and September 2021, 510 patients with confirmed HBV infection based on the presence of HBsAg tested with a commercially available enzyme-linked immunosorbent assay (ELISA) kit were referred to the Gastroenterology and Liver Research Clinic affiliated with Shiraz University of Medical Sciences, Shiraz, Iran. Based on clinical and laboratory data, 83 patients were classified as chronically infected with HBV and were positive for HBsAg for over 6 months. Of the 83 patients with chronic HBV infection, 24 and 59 cases were female and male, aged 16-60 (34.12 ± 12.2) and 16 - 70 (37.11 ± 12.46) years, respectively. All the patients provided written informed consent before participating in the study. The inclusion criteria for study participants were the coexistence of HBsAg and anti-HBs and a negative result for other viral markers, such as hepatitis C virus and human immunodeficiency virus.
Five milliliters of peripheral blood was collected from each patient in a sterile tube. Serum samples were obtained by the centrifugation of the blood, and the serum was divided into aliquots and stored at - 20°C until use. The blood samples to detect HBV DNA were collected in separate tubes to avoid cross-contamination to minimize contamination from other subjects’ samples.
Figure 1 shows the STROBE flowchart for patient selection.
3.2. Serologic Testing
Serological markers for HBV infection, including HBsAg, anti-HBs, anti-HBc, HBeAg, and anti-HBe, were analyzed using commercially available kits (Dia.Pro ELISA kit, Italy) according to the manufacturer’s instructions. Serum alanine aminotransferase levels were also measured using automated techniques. The quantitative determination of the marker concentration was calibrated and taken into account according to the criteria of the Dia.Pro kit.
3.3. DNA Extraction and Polymerase Chain Reaction Assay
Viral DNA was extracted from 100 μL serum using the DNP kit (Fermentas, Italy) according to the manufacturer’s instructions. First, a polymerase chain reaction (PCR) assay was performed to detect and confirm the presence of HBV DNA in clinical specimens using the commercially available kit. For this purpose, the HBV PCR detection kit (Cat. No. PR783IC, CinnaGen, Tehran, Iran) was used to amplify the 353 bp DNA fragment of the highly conserved region of the S gene. Then, following the manufacturer’s instructions, a 15 μL reaction mixture containing 10 μL template DNA and 2.0 units of Taq DNA polymerase was subjected to two rounds of PCR amplification. Amplification was performed using the following program:
One cycle of 93°C for 2 minutes, 61°C for 20 seconds, and 72°C for 40 seconds, followed by 35 cycles of 93°C for 20 seconds, 61°C for 20 seconds, and 72°C for 40 seconds, followed by 5 minutes extension at 72°C.
Electrophoresis on gel agarose was performed for the last diagnosis of produced DNA fragments (353 bp).
3.4. Amplification of HBV Surface Gene (S) and Sequence Analysis
The HBV surface (S) gene was amplified by a nested PCR, using the outer forward primers HB1F (forward) 5´-AAG CTC TGC TAG ATC CCA GAG T-3’/ HB2R (reverse): 5´-CAT ACT TTC CAA TCA ATA GG-3’ for the first round (972 bp) and inner primers forward primer P7 5´-GTG GTG GAC TTC TCT CAA TTT TC-3’ and inner reverse primer P8 5′-CGG TAW AAA GGG ACT CAM GAT-3 for the second round (541 bp).
The first-round PCR was performed for 35 cycles (94°C for 40 seconds, 55°C for 40 seconds, and 72°C for 1 minute), and a final extension step was performed at 72°C for 4 minutes in a 25 μL reaction volume. The reaction mixture contained 50 ng of extracted DNA, 1X PCR buffer, 200 mM of each deoxynucleoside triphosphate, 1.5 mM MgCl2, 10 pmol/mL of each outer primer, and 1 unit of Taq DNA polymerase (Invitrogen, France). Then, 1 microliter of the first-round PCR product was subjected to the second-round PCR under the same conditions but with internal primers.
Appropriate positive and negative controls were included in each assay. After the second round, a 541 bp fragment was obtained and detected by electrophoresis in a 2% agarose gel. The PCR product was purified using a DNA purification kit (New England Biolabs, Frankfurt, Germany) and bidirectionally sequenced with internal primers (SeqLab, Germany). Wild-type HBV PreS1, PreS2, and S sequences, genotype D, and PCR product sequences were determined and converted to related amino acids (protein BLAST, the National Center for Biotechnology Information [NCBI]) for further analysis.
3.5. Phylogenetic Analysis and Sequence Alignment
In this study, the nucleotide sequences of HBV isolates were aligned using ClustalW2 software (https://www.ebi.ac.uk/Tools/msa/clustalw2/) with reference sequences from Genebank, which had accession numbers AY653847, AY233293, and EU414137. Additionally, the phylogenetic trees were created using the same software.
3.6. Determination of Hydropathy Index for Amino Acid Residues in MHR
The hydropathic index of each amino acid is a number that indicates the degree of hydrophobicity or hydrophilicity of its side chains. The higher the hydropathy index, the more hydrophobic the amino acid is. In the protein structure, the hydrophobic amino acids must tend to be within the third structure of the protein. On the other hand, hydrophilic amino acids are mostly found on the protein’s surface.
Using the online software ProtScale and EpiQuest® (version 4.0.0.1), all changes in the amino acid residues of the HBsAg were analyzed to determine whether mutations could affect the structure of the epitope and the appearance of an escape mutant.
4. Results
4.1. Serological Results
Among 83 patients with chronic HBV infection, the coexistence of HBsAg and anti-HBs were observed in 11 individuals (13.25%) (Table 1).
| No. | Age | Gender | HBsAg | HBsAb mIU/mL | HBeAg | HBeAb | AST | ALT | Lamivudine |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 37 | M | + | 18.65 (+) | ND | ND | 15 | 18 | NR |
| 2 | 35 | M | + | 26.82 (+) | ND | ND | 34 | 82 | NR |
| 3 | 21 | M | + | 13.69 (+) | ND | ND | 45 | 68 | NR |
| 4 | 16 | F | + | 29.64 (+) | ND | + | 46 | 105 | NR |
| 5 | 27 | M | + | 10.19 (+) | ND | ND | 130 | 380 | NR |
| 6 | 46 | M | + | 12.14 (+) | - | + | 22 | 31 | NR |
| 7 | 56 | M | + | 19.82 (+) | - | + | 39 | 38 | NR |
| 8 | 45 | M | + | 10.68 (+) | + | + | 78 | 93 | NR |
| 9 | 34 | M | + | 10.07 (+) | + | + | 33 | 25 | R |
| 10 | 52 | M | + | 17.70 (+) | - | + | 208 | 380 | R |
| 11 | 20 | M | + | > 250 (+) | - | + | 51 | 68 | R |
Abbreviations: HBsAg, hepatitis B surface antigen; HBsAb, hepatitis B virus surface antibody; AST, aspartate aminotransferase; ALT, alanine aminotransferase; R, received; NR, not received; F, female; M, male; ND, not done.
a ALT normal < 41 U/L, AST normal < 31 U/L.
4.2. Detection of DNA in Serum Samples
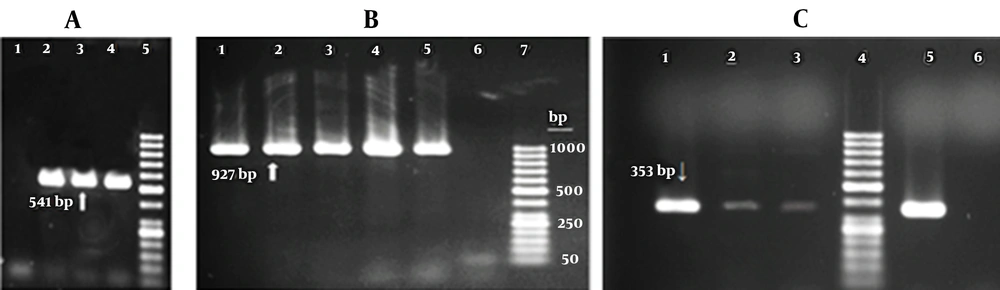
All 11 serum samples from subjects with positive HBsAg and anti-HBs were positive for HBV DNA using primers to amplify the full-length HBsAg gene (927 bp) or nested PCR primers to amplify MHR “a” determinant of HBsAg (541 bp). Figure 2 shows the HBV DNA results of some patients. Figure 2C illustrates HBV DNA detection in the first screening PCR assay.
Agarose gel electrophoresis of DNA extracted from serum samples of patients with the coexistence of hepatitis B surface antigen (HBsAg) and Hepatitis B Virus surface antibodies. A, Results of nested polymerase chain reaction (PCR) for Amplification of major hydrophilic region (1: Negative control, 2 - 4: Patients’ serum samples, 5: 50 bp DNA ladder); B, Results of PCR amplification using primers for the complete HBsAg sequence (1 - 5: Patients’ serum samples, 6: Negative control, 7: 50 bp DNA ladder); C, Results of first screening PCR for detection of hepatitis B virus DNA in serum samples of three patients (1 - 3: Patients’ serum samples, 5: Positive control, 6: Negative control)
4.3. DNA Sequence Analysis and Hydropathy Analysis
It was possible to obtain nucleotide sequence results for 11 cases; however, only seven of them were thoroughly analyzed. The NCBI reference sequences AY653847, EU414137, and AY233293 from GeneBank were used to compare the PCR product nucleotide sequences. Multiple sequence alignments and phylogenetic analysis of HBV S gene sequences from the seven patients were conducted using ClustalW2 software (https://www.ebi.ac.uk/Tools/msa/clustalw2/) to identify clusters and assess the epidemiologic features of infections. However, due to the limited sample size and suboptimal visual quality of the phylogenetic tree, the results of the phylogenetic analysis are presented in the supplementary materials (Appendix). The phylogenetic analysis revealed that the genotype of HBV observed in the studied region belonged to genotype D, which includes viruses circulating in Eurasia.
In the next step, the Expasy: Swiss Bioinformatics Resource Portal was used to convert the sequence of the identified PCR product to the corresponding amino acid. The sequences were then specifically compared to wild type and HBV genotype D.
ProtScale online software was used to deeply predict surface protein hydrophilicity and hydrophobicity. The hydropathy index of an amino acid is a number that indicates the hydrophobic or hydrophilic properties of its side chain. Antigenic plot and antigenic determinants were demonstrated for each sequence using online EpiQuest® software (version 4.0.0.1) and compared to wild type to find the effect of sequence changes on the hydropathy index.
Based on the above-mentioned theory and with the bioinformatics program, an antigenic diagram was drawn for each sequence, and the antigenic determinants were determined. Then, by comparing the antigenic determinants of each sequence to the antigenic determinants of the wild type, it was determined whether or not the disclosed changes in each sequence represented a change in the hydropathy index of the amino acids and consequently, a difference in the degree of hydrophobicity or hydrophilicity of their side chains. The results showed that all mutations were upstream and downstream of the MHR. However, there was a one-point mutation within the MHR of HBsAg, as shown in Table 2.
| Sample | Amino Acid Position of Wild Type to Mutant | ||
|---|---|---|---|
| MHR Upstream Mutations | MHR Downstream Mutations | Mutation Within MHR | |
| 1 | S64L | I198T | R207S |
| 2 | I189T, Y206F | R207S | |
| 3 | Q54 R | I189T | R207S |
| 4 | I189T, V190G | Y134F, R207S | |
| 5 | I189T | R207S | |
| 6 | I189T, Y206F | R207S | |
| 7 | I189T | R207S | |
Abbreviations: MHR, major hydrophilic region; F, phenylalanine; G, glycine; I, isoleucine; L, leucine; Q, glutamine; R, arginine; S, serine; T, threonine; V, valine; Y, tyrosine.
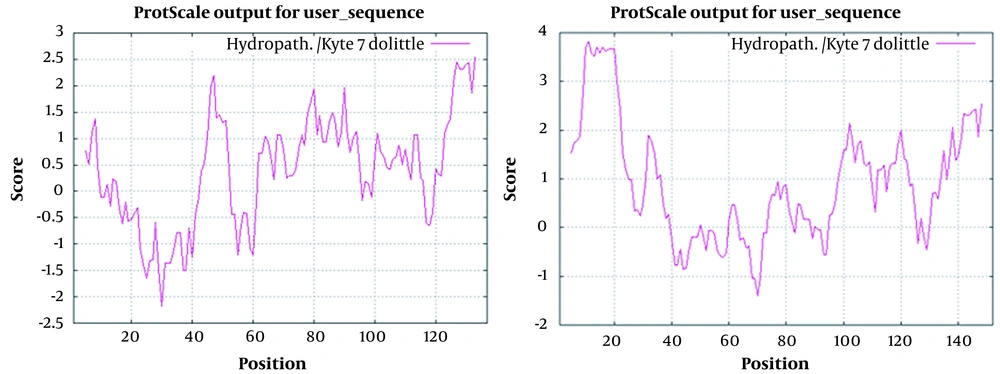
The results showed that replacing hydrophilic amino acids with hydrophobic amino acids and vice versa created new variants. In addition, the third structure of the epitope was also changed. For example, in sample 4, the hydropathic plot and antigenic determinants were altered due to mutation in four different positions within and downstream of the MHR (Figures 3 and 4). Consequently, the structures of the amino acids also changed, resulting in an altered shape of the epitopes (Figure 4).
Left: Hydropathy Plot for Small Hepatitis B Surface Antigen (HBsAg) of Patient No. 4
Right: Hydropathy Plot for Small HBsAg with Accession Number GQ183486 (Nearest HBsAg Similar to Iranian Isolates)
This study examined the HBsAg hydropathic profile and epitope predictions using the Immune Epitope Database Analysis Resource (http://tools.iedb.org/main/). No results were obtained when T-cell epitope prediction tools were used. This set of tools includes major histocompatibility complex class I and II binding predictions. However, B-cell epitope prediction tools predicted linear B-cell epitopes based on the sequence characteristics of the antigen using amino acid scales (Figure 5).
Epitope prediction of hepatitis B surface antigen (HBsAg) isolated from patient No. 4 in comparison to the reference HBsAg with accession number GQ183486 (Nearest HBsAg Similar to Iranian Isolates). Right: Hydropathy plot for small HBsAg with accession number GQ183486 (Nearest HBsAg similar to Iranian isolates). Left: Hydropathy plot for small HBsAg of patient No. 4
5. Discussion
Although most escape mutants in HBV infection are caused by changes in the “a” determinant and MHR, mutations that occur outside this region can result in the emergence of mutants that are resistant to the immune system and have great resistance (7, 10). The HBsAg and anti-HBs might generally coincide because a mutation in the S gene might occur naturally or after treatment. Additionally, the existence of different genotypes and subtypes in one patient and the high titer of anti-HBs might affect the emergence of an HBV escape mutant in a patient (19).
The present study investigated the clinical and virological characteristics of patients with coexisting HBsAg and anti-HBs in Fars province, Iran. It was shown that of 83 patients with a definitive diagnosis of chronic HBV infection, 11 subjects (13.2%) were simultaneously positive for anti-HBs and HBsAg.
The tested samples in the current study were the HBV genotype D and adw subtype. In addition, all but 1 of these 11 serum samples had an anti-HBs level of less than 100 mIU/mL. Therefore, there is a possibility that the low anti-HBs level (anti-HBs < 100 mIU/mL) in these serum samples does not bind to HBsAg in patients with chronic hepatitis. Mutations in eight amino acids of seven samples analyzed for nucleotide sequencing were found at 27 different sites in three locations, namely upstream, within, and downstream of the MHR (Table 2).
The humoral immune response to S-region proteins on the surface antigen has been shown to result in the clearance of the virus from peripheral blood (7, 20). On the other hand, the cellular immune response eliminates liver cells infected with the virus. Consequently, any changes in the S region (i.e., in the MHR associated with HBsAg) give rise to HBV types termed “escape mutants”. As a result, these viruses can evade the immune system. This phenomenon leads to a stable state of the virus or progression of HBV infection (21).
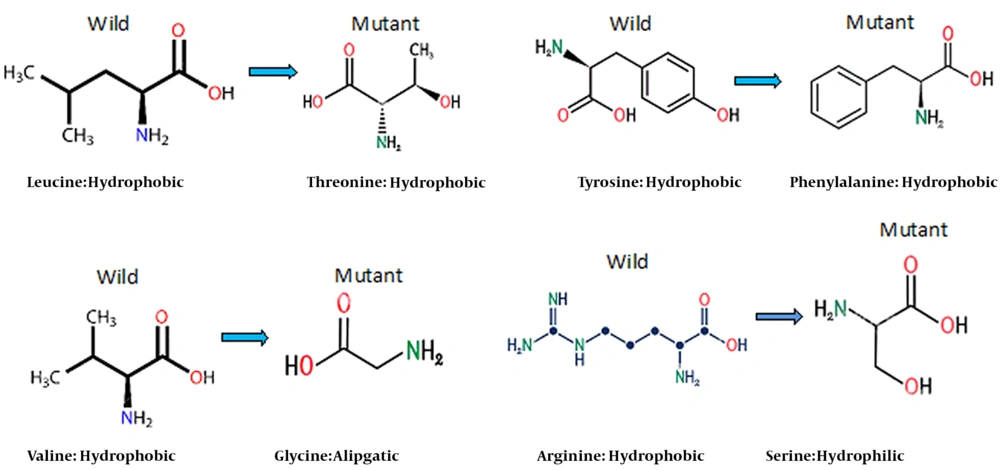
The replacement of the amino acid (S) serine, which is a hydrophilic amino acid with one carbon molecule, with the amino acid (L) leucine, with four carbon molecules, which is hydrophobic, S64L, upstream the MHR protein, can change the shape of the protein molecule. In addition, the change leads to alterations in the reaction of the HBsAg with the antibody. However, another mutation occurred in the same region, Q54R, where two hydrophilic amino acids were displaced. Since these amino acids also differ in their molecular structure, it is still possible to affect the MHR protein in antigen-antibody or antigen-antigen reactions with immune systems.
In the seven samples, the amino acid isoleucine (I), V152I, was substituted for valine (V) in amino acid #152, both of which are hydrophilic amino acids. This change did not affect the epitope as both are similar in isomeric amino acid structure. The presence of an extra-base (-CH3) in isoleucine is the only difference between these two amino acids. However, another change involves changing the amino acid tyrosine (Y), an amphoteric amino acid, with amino acid phenylalanine (F) at position 137 of the MHR, Y137F, in a sample. These two amino acids are very similar in structure; however, tyrosine is a polar, uncharged amino acid; nevertheless, phenylalanine is a non-polar amino acid. Therefore, this shift can also alter the epitope structure of the MHR.
Another difference was substituting the amphoteric amino acid tyrosine (Y) for the phenylalanine (F) at position 137 of the MHR, Y137F, in a sample. The structures of these two amino acids are very similar; however, tyrosine is a polar amino acid with no charge. In contrast, phenylalanine is a non-polar amino acid, and this is a polar amino acid. This shift can also alter the epitope structure of the MHR.
The antigenic determinant “a” is related to the antigen-antibody reaction, and several escape mutations are associated with it (22). Since only one case had a mutation in the determining region “a”, the coexistence of HBsAg and anti-HBs in patients with chronic hepatitis is not necessarily linked to the change in this region. On the other hand, the changes that have occurred in the two upstream and downstream areas of this region can, to some extent, justify the simultaneous presence of HBsAg and anti-HBs in these patients with chronic hepatitis. Many of the mutations in these samples were downstream of the MHR. Among the seven cases for which the sequence determination test was performed, in all seven patients, genetic changes were observed in the downstream region related to the MHR. These changes include I189T, V190G, Y206F, and R207S.
Previous studies have shown that mutation at amino acid numbers 184 to 216 and 45 to 79 can be a major factor in developing HCC in patients (23). Increasing antiviral agents against HBV polymerase can produce new mutations in HBsAg molecules. Mutations caused by drug use are often located downstream of the MHR (24). Although these changes have been studied to some extent, changes in amino acids 206, 207, 189, and 190 in the current investigated patients might also indicate a similar effect since all of these reactions occurred downstream of the MHR, which is also associated with antigen and antibody response. In this study, a patient receiving the antiviral drug lamivudine developed a mutation in the MHR. Because lamivudine can cause a mutation in the HBV polymerase gene that overlaps with HBsAg, the mutation could also occur in this manner.
The Immune Epitope Database Analysis Resource (http://tools.iedb.org/main/) was used to examine HBsAg hydropathy profiles and epitope predictions. As shown in Figure 5, there are some changes in the epitope sequences and some minor changes in the phenotype of the antigen. However, there is no evidence that these changes can affect antigen-antibody responses. Based on this analysis, no significant changes were observed in the Th and CTL immune epitopes. However, nucleotide changes might affect B-cell-dependent epitopes; nevertheless, the present study did not investigate these effects.
Overall, the current study’s results suggest that the coexistence of HBsAg and anti-HBs is not solely due to an amino acid substitution in the “a” determinant. Nevertheless, the mutation downstream and upstream of the MHR plays a major role in this coexistence. Another reason for the coexistence of HBsAg and anti-HBs is a low anti-HBs titer (< 100 mIU/mL) in patients with chronic hepatitis who cannot bind to HBsAg.