1. Background
Non-alcoholic fatty liver disease (NAFLD) is a metabolic disorder leading to the abnormal accumulation of fat in the liver (1). Some studies demonstrated the high (> 20%) prevalence of NAFLD among adult general populations around the world (2-4). The non-symptomatic NAFLD can progress to non-alcoholic steatohepatitis (NASH) and, in the long term, to advanced liver diseases such as cirrhosis and hepatocellular carcinoma (HCC) (5). There is little success with the development of therapeutic agents for NAFLD and its advanced complications; however, lifestyle modification in terms of diet and exercise to weight loss can prevent the progression of NAFLD or even cause its regression (6).
A wide spectrum of modifiers can contribute to development of NAFLD or its progression to advanced liver diseases (7). These include genetic predisposition, age, lifestyle, diabetes, etc. (7). Recently, a great interest has emerged in the elucidation of the role of genetic parameters in the pathogenesis of NAFLD (8). In this context, the genome-wide association studies (GWASs) contributed greatly to finding new genetic variations connected with NAFLD, including PNPLA3 rs738409, TM6SF2 rs58542926, GCKR rs1260326, etc. (8). The PNPLA3 rs738409 G allele produces an altered variant known as I148M (Ile148Met), which would fail fat regulation and leads to increased production and decreased breakdown of fats in the liver (9). The TM6SF2 rs58542926 T allele forms a missense variant called K167E (Lys167Glu), which is associated with reduced triglyceride-rich lipoproteins secretion into the serum and an excessive lipid accumulation, triggering NAFLD (10). Following the GWASs, the associated SNPs were tested mostly in clinical settings in different ethnic populations to confirm the GWAS findings. In these clinical studies, PNPLA3 rs738409 was found to be an important player in NAFLD pathogenesis, while the studies on TM6SF2 rs58542926 were not definitive. Although most clinic-based studies confirmed the association of PNPLA3 rs738409 with NAFLD, there are scarce pieces of evidence on the role of this SNP in NAFLD cases through population-based recruitment (11-13). Moreover, there are no reliable pieces of evidence on the association of PNPLA3 and TM6SF2 polymorphisms with NAFLD in the Iranian population.
2. Objectives
This study aimed to assess the association of PNPLA3 rs738409 and TM6SF2 rs58542926 SNPs with NAFLD in two study recruitment settings of population-based NAFLD (PB-NAFLD) and clinic-based NAFLD (CB-NAFLD) participants.
3. Methods
3.1. Study Design and Study Participants
This is a case-control study in two different groups of NAFLD participants through population-based and clinic-based recruitment. The population-based recruitment was based on a population-based study previously conducted in Shiraz (south of Iran) in 2010 - 2011 (2). The PB-NAFLD participants and age- and sex-matched population-based healthy individuals without NAFLD (control group) were sampled from participants of the above-mentioned population-based study. The participants of the original population-based study were randomly selected adult (> 18 years) residents of seven municipality regions of Shiraz city. The clinic-based cohort was recruited from the Middle East Liver Diseases (MELD) Center, a tertiary liver clinic in Tehran, Iran. The patients with NAFLD referred to the MELD Center mostly consisted of those with NAFLD-related liver diseases, such as NASH, high liver fibrosis, cirrhosis, HCC, and the extrahepatic manifestation of liver diseases. The CB-NAFLD participants were recruited consecutively from adult (> 18 years) patients who attended the MELD Center in 2018 - 2019. The diagnosis of NAFLD in both cohorts was based on the transabdominal ultrasonography modality performed by experienced radiologists. In both cohorts, participants with current hepatitis B or hepatitis C infections, autoimmune liver diseases, Wilson’s disease, hemochromatosis, alcohol consumption (> 40 g/day for males and > 20 g/day for females), and pregnancy were excluded. The study protocol of the population-based study was previously approved by the Research Ethics Committee of the Health Policy Research Center affiliated with Shiraz University of Medical Sciences. The study protocol of the clinic-based recruitment was approved by the Research Ethics Committee of the Islamic Azad Tehran Medical Sciences University, Pharmacy and Pharmaceutical Branches Faculty, with approval identification code of IR.IAU.PS.REC.1397.104. All participants provided written informed consent including those provided in the original population-based study. The study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki.
Data collection in the population-based study was based on a questionnaire-based interview, physical examinations, and laboratory testing in the context of research. Data collection in the clinic-based study was based on the clinical management of NAFLD collected as clinical and laboratory data in the patients’ electronic records of the MELD Center.
3.2. Genotyping of PNPLA3 and TM6SF2 SNPs
The buffy coat was separated from the blood specimens of all study participants and kept frozen at -20°C until further assessments. Genomic DNA was extracted using the QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany) aligned with the manufacturer’s instructions. In this study, PNPLA3 rs738409 and TM6SF2 rs58542926 SNPs were genotyped using the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method. Primer design for PCR-RFLP was performed using Primer-BLAST software, and the specificity of primers was evaluated using BLAST software (14). The designed primers for both PNPLA3 rs738409 (PNP-F & PNP-R) and TM6SF2 rs58542926 (TM-F & TM-R) are listed in Appendix 1. The PCR was performed using ready-to-use Taq Polymerase Master Mix RED (Ampliqon, Denmark) according to the manufacturer’s instructions. To perform PCR-amplification in a 20-µL reaction, 100 - 500 ng of the extracted genomic DNA was amplified using 0.5 µM of each forward and reverse primers for both polymorphisms in separate reactions. The PCR temperature profile for both PNPLA3 and TM6SF2 reactions were the same as follows: 94°C for 5 minutes, 40 cycles of 94°C for 20 seconds, 65°C for 20 seconds, and 72°C for 20 seconds, followed by 4°C for 10 minutes. For the RFLP analysis, the PCR products of PNPLA3 rs738409 (291 bp) and TM6SF2 rs58542926 (249 bp) were digested with 10 units of BseGI and HPY188I overnight, respectively. Digested PCR products were electrophoresed on 3% agarose gel and 100 bp DNA Ladder was electrophoresed alongside digested PCR products. Digestion of PNPLA3 rs738409 291 bp PCR product showed one undigested 291 bp fragment for GG genotype, two fragments of 225 and 66 bp in CC genotype, and three fragments of 225, 66, and 291 bp in CG genotype, while the PCR-RFLP pattern for TM6SF2 rs58542926 showed two fragments of 177 and 72 bp in the CC genotype and three fragments of 177, 72, and 249 bp in the CT genotype.
For validation of the PCR-RFLP method, 10 samples for each of the PNPLA3 rs738409 and TM6SF2 rs58542926 genotypes were re-evaluated using the DNA sequencing method.
3.3. Statistical Analysis
The collected data were entered into a dataset and analyzed using IBM SPSS version 22. The categorical variables were presented as numbers and percentages, and the numerical variables as mean and standard deviation (SD) for those with normal distribution, and the median and interquartile range (IQR) for variables that deviated from the normal distribution. Hardy-Weinberg Equilibrium (HWE) was calculated for both SNPs using Excel software. One-way ANOVA and Kruskal-Wallis tests were used to compare the numerical variables with a normal distribution and with deviation from a normal distribution, respectively, followed by multiple comparison tests to compare the mean/median between the three study groups. Pearson chi-square and Fisher's exact tests compared the categorical variables. For multivariate analysis, NAFLD modifiers with P < 0.1 were entered into the binary logistic regression model. Excel software was used for the construction of forest plots. P < 0.05 was considered statistically significant.
4. Results
4.1. Characteristics of the Study Population
A total of 110 PB-NAFLD and 110 sex- and age-matched healthy individuals were selected from the population-based study. An additional 73 NAFLD cases were recruited from the MELD center as CB-NAFLD participants. Genotyping of SNPs was successful in all samples except two PB-NAFLD participants, which failed for both SNPs resulting in the inclusion of 108 PB-NAFLD participants.
Regarding the characteristics of the study population, most of the participants were middle-aged individuals and the CB-NAFLD participants were significantly older than people in the PB-NAFLD and control groups. In all three groups, more than half of the participants were males, with a slightly higher proportion of male sex in the CB-NAFLD group (Table 1). Body mass index (BMI), fasting plasma glucose (FPG), alanine transaminase (ALT), and aspartate transaminase (AST) were significantly higher in both NAFLD groups than in the control group; however, it was not significantly different between the two NAFLD groups. Triglyceride level was significantly higher in the PB-NAFLD participants than in the control and CB-NAFLD participants. Cholesterol was significantly different among the groups and was the highest in the PB-NAFLD group, followed by the control group. The Aspartate Aminotransferase to platelet ratio index (APRI) was significantly higher in the CB-NAFLD group than in the two other groups. The frequency of diabetes mellitus, hypertension, hyperlipidemia, and metabolic syndrome was significantly higher in the PB-NAFLD group than in the control group (Table 1). Among the CB-NAFLD participants, 26.8% had cirrhosis. The detailed characteristics of the three study groups are presented in Table 1.
| Characteristics | Control (n = 110) | Population-Based NAFLD (n = 108) | Clinic-Based NAFLD (n = 73) | P-Value |
|---|---|---|---|---|
| Age (y) | 47.6 ± 10.9 | 47.6 ± 11.2 | 52.0 ± 12.3 * | 0.02 d |
| Sex | 0.69 e | |||
| Male | 61 (55.5) | 61 (56.5) | 45 (61.6) | |
| Female | 49 (44.5) | 47 (43.5) | 28 (38.4) | |
| BMIf | 25.0 ± 3.9 * | 28.9 ± 4.3 | 29.6 ± 4.3 | < 0.01 d |
| FPGf (mg/dL), median (IQR) | 84 (16) * | 97 (28) | 97.5 (26) | < 0.01 g |
| TGf(mg/dL), median (IQR) | 119 (54) | 161 (107) * | 132.5 (135) | < 0.01 g |
| Cholesterolf(mg/dL), median (IQR) | 192 (54) ** | 200 (63) ** | 173 (55) ** | < 0.01 g |
| ALTf (IU/L), median (IQR) | 20 (13) * | 29 (19) | 32.5 (39) | < 0.01 g |
| ASTf(IU/L), median (IQR) | 22 (8) * | 27 (13) | 32 (22) | < 0.01 g |
| APRIf, median (IQR) | 0.2 (0.1) | 0.3 (0.2) | 0.4 (0.5) * | < 0.01 g |
| Diabetes mellitusf | < 0.01 h | |||
| Yes | 3 (2.8) | 15 (14.2) | NA | |
| No | 106 (97.2) | 91 (85.8) | NA | |
| Hypertensionf | 0.03 h | |||
| Yes | 10 (9.2) | 21 (19.8) | NA | |
| No | 99 (90.8) | 85 (80.2) | NA | |
| Hyperlipidemiaf | < 0.01 h | |||
| Yes | 17 (15.6) | 33 (31.1) | NA | |
| No | 92 (84.4) | 73 (68.9) | NA | |
| Metabolic syndrome | < 0.01 h | |||
| Yes | 14 (12.7) | 43 (39.8) | NA | |
| No | 96 (87.3) | 65 (60.2) | NA | |
| Cirrhosisf | - | |||
| Yes | NA | NA | 19 (26.8) | |
| No | NA | NA | 52 (73.2) |
Abbreviations: NAFLD, non-alcoholic fatty liver disease; n, number; BMI, body mass index; FPG, fasting plasma glucose; IQR, interquartile range; TG, triglycerides; ALT, alanine aminotransferase; AST, aspartate aminotransferase; APRI, aspartate aminotransferase to platelet ratio index; NA, not available.
a Values are expressed as No. (%) or mean ± SD unless otherwise indicated.
b * Following multiple comparisons, the value is significantly different between the defined group and other groups.
c ** Following multiple comparisons, the value is significantly different across the defined groups.
d One-way ANOVA.
e Pearson chi-Square.
f There was less than 5% missing data.
g Kruskal-Wallis test.
h Fisher's exact test.
4.2. Association of PNPLA3 and TM6SF2 with Non-alcoholic Fatty Liver Disease
The results of DNA sequencing of 10 rs738409 CC, 10 rs738409 CG, 10 rs738409 GG, 10 rs58542926 CC, and 10 rs58542926 CT samples were 100% concordant with the PCR-RFLP results, confirming the whole PCR-RFLP genotyping process. The HWE based on the distribution of genotypes was assessed in the whole study population and each study group for both PNPLA3 rs738409 and TM6SF2 rs58542926 SNPs. The distribution of both SNPs in all study groups was in HWE (Appendix 2).
The distribution of PNPLA3 rs738409 genotypes (genotypic model), alleles (allelic model), recessive genotype GG (recessive model), and dominant genotype CC (dominant model) was not significantly different between the PB-NAFLD and control groups (Table 2). Moreover, the distribution of PNPLA3 rs738409 genotypes (genotypic model), alleles (allelic model), and dominant genotype CC (dominant model) was not significantly different between the CB-NAFLD and control groups; however, the recessive genotype GG (recessive model) had more frequency in the CB-NAFLD group than in the control group (19.2% vs. 8.2%; P = 0.04, OR = 2.66, 95% CI: 1.09 - 6.54) (Table 2). In an additional analysis, the distribution of PNPLA3 rs738409 genotypes was significantly different between the PB-NAFLD and CB-NAFLD groups in the genotypic, allelic, dominant, and recessive models (P = 0.02, P < 0.01, P = 0.03, and P = 0.02, respectively).
| Genetic Model and rs738409 Genotypes/Alleles | Population-Based NAFLD, No. (%) | Clinic-Based NAFLD, No. (%) | Control, No. (%) | PB-NAFLD vs. Control | CB-NAFLD vs. Control | ||
|---|---|---|---|---|---|---|---|
| OR (95% CI) | P-Value a | OR (95% CI) | P-Value a | ||||
| Genotypic model | |||||||
| CC | 65 (60.2) | 32 (43.8) | 52 (47.3) | 1 | 1 | ||
| CG | 36 (33.3) | 27 (37.0) | 49 (44.5) | 0.59 (0.33 - 1.03) | 0.09 | 0.90 (0.47 - 1.70) | 0.75 |
| GG | 7 (6.5) | 14 (19.2) | 9 (8.2) | 0.62 (0.22 - 1.78) | 0.43 | 2.53 (0.98 - 6.49) | 0.06 |
| Recessive model | |||||||
| CC+CG | 101 (93.5) | 59 (80.8) | 101 (91.8) | 1 | 1 | ||
| GG | 7 (6.5) | 14 (19.2) | 9 (8.2) | 0.78 (0.28 - 2.17) | 0.80 | 2.66 (1.09 - 6.54) | 0.04 |
| Dominant model | |||||||
| CC | 65 (60.2) | 32 (43.8) | 52 (47.3) | 1 | 1 | ||
| GG+CG | 43 (39.8) | 41 (56.2) | 58 (52.7) | 0.59 (0.35 - 1.02) | 0.06 | 1.15 (0.63 - 2.08) | 0.65 |
| Allelic model | |||||||
| C | 166 (76.9) | 91 (62.3) | 153 (69.5) | 1 | 1 | ||
| G | 50 (23.1) | 55 (37.7) | 67 (30.5) | 0.69 (0.45 - 1.05) | 0.10 | 1.38 (0.89 - 2.15) | 0.17 |
Abbreviations: NAFLD, non-alcoholic fatty liver disease; No., number; PB, population-based; CB, clinic-based; OR, odds ratio; CI, confidence interval.
a Fisher's exact test.
The distribution of TM6SF2 rs58542926 genotypes (genotypic model) and alleles (allelic model) was not significantly different between the PB-NAFLD and control groups (Table 3). Besides, the distribution of TM6SF2 rs58542926 genotypes (genotypic model) and alleles (allelic model) was not significantly different between the CB-NAFLD and control groups (Table 3).
| Genetic Model and rs58542926 Genotypes/Alleles | Population-Based NAFLD, No. (%) | Clinic-Based NAFLD, No. (%) | Control, No. (%) | PB-NAFLD vs. Control | CB-NAFLD vs. Control | ||
|---|---|---|---|---|---|---|---|
| OR (95% CI) | P Value a | OR (95% CI) | P Value a | ||||
| Genotypic model | |||||||
| CC | 97 (89.8) | 63 (86.3) | 101 (91.8) | 1 | 1 | ||
| CT | 11 (10.2) | 10 (13.7) | 9 (8.2) | 1.27 (0.51 - 3.21) | 0.65 | 1.78 (0.69 - 4.62) | 0.32 |
| Allelic model | |||||||
| C | 205 (94.9) | 136 (93.2) | 211 (95.9) | 1 | 1 | ||
| T | 11 (5.1) | 10 (6.8) | 9 (4.1) | 1.26 (0.51 - 3.10) | 0.65 | 1.72 (0.68 - 4.35) | 0.34 |
Abbreviations: NAFLD, non-alcoholic fatty liver disease; No., number; PB, population-based; CB, clinic-based; OR, odds ratio; CI, confidence interval.
a Fisher's exact test
4.3. Multivariate Analysis of Non-alcoholic Fatty Liver Disease Modifiers
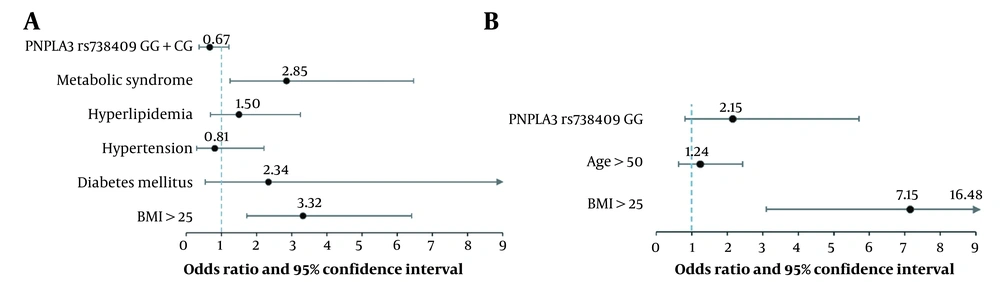
In the multivariate analysis, metabolic syndrome (OR = 2.85) and BMI > 25 (OR = 3.32) remained as the independent modifiers of NAFLD in the PB-NAFLD cohort (Figure 1A). In addition, BMI > 25 (OR = 7.15) remained as a single independent modifier of NAFLD in the CB-NAFLD cohort (Figure 1B).
Multivariate analysis of modifiers of non-alcoholic fatty liver disease (NAFLD). A, Regression model including PNPLA3 rs738409 SNP (dominant model), metabolic syndrome, hyperlipidemia, hypertension, diabetes mellitus, and BMI in the evaluation of population-based NAFLD participants; B, Regression model including PNPLA3 rs738409 SNP (recessive model), age, and BMI in the evaluation of clinic-based NAFLD participants.
5. Discussion
This study included two groups of individuals with NAFLD, i.e., PB-NAFLD and CB-NAFLD, to trace the association of globally most-studied genetic variants with NAFLD in the Iranian population. The PB-NAFLD group consisted of individuals with a study-driven diagnosis of NAFLD, mostly unaware of the presence of liver steatosis, while the CB-NAFLD consisted of patients referred to a tertiary liver clinic for the management of NAFLD-related liver diseases. This is also evident from the APRI data with a significantly higher APRI value in the CB-NAFLD than in PB-NAFLD. This represented two distinct groups of NAFLD for studying the genetics parameters. Globally, most studies on the association of genetic variations with NAFLD recruited their participants from clinics, indicating that the included NAFLD cases were those mostly with liver diseases rather than those with only steatosis and without inflammation or fibrosis (15, 16). In studies with the recruitment of NAFLD cases with the dominance of NASH and liver fibrosis, it is obscured whether the genetic variations such as PNPLA3 rs738409 contributed to the initial development of steatosis or progression of steatohepatitis/liver fibrosis or both while studies including NAFLD cases from the general population would mostly elucidate the role of genetic markers in the initial development of steatosis, especially with subgroup analysis by steatosis and liver fibrosis degrees (17, 18).
The role of PNPLA3 rs738409 has been evaluated in many studies throughout the world. Most of the studies have found an association between this SNP and NAFLD (19, 20). The current study found no association between PNPLA3 rs738409 and NAFLD in the PB-NAFLD while there was a significant association between this polymorphism and NAFLD in the CB-NAFLD. Based on the findings of the current study, we postulate the prominent role of PNPLA3 rs738409 in the progression of the liver disease caused by NAFLD; however, the tentative role of PNPLA3 rs738409 in the initial development of NAFLD was not observed in the current study. A recently published GWAS with multiethnic population-based recruitment found PNPLA3 rs738409 to be strongly associated (P < 1 × 105) with Percent Liver Fat only in Japanese-Americans and European-Americans while Latinos and African-Americans reached a P < 0.05 and Native Hawaiians showed no statistically significant association (P = 0.101) (21). The latter variation in the ethnicity-specific observation can be caused by the population-based sampling and the lower magnitude of rs738409 in the development of steatosis than that for the progression of liver diseases, globally observed in the clinical studies. The relationship between PNPLA3 rs738409 and NAFLD had not been well presented in the Iranian population in previous studies. Just there are two case-control studies considering the PNPLA3 rs738409 genotypes distribution in Iranian individuals with NAFLD (22, 23). Surprisingly, both studies found the C allele (I variant) as the risk allele, which seems to be caused by an erroneous interpretation of the genotyping results.
Regarding TM6SF2 rs58542926, the association of the K167E variant with NAFLD and NAFLD-related liver diseases was studied previously (24, 25). These studies confirmed the modest association of TM6SF2 rs58542926 with NAFLD. However, we found no association between this polymorphism and NAFLD in the Iranian population. In line with our findings, Lisboa et al. (15) did not find the TM6SF2 rs58542926 polymorphism as a modifier of NAFLD in the Brazilian population.
In the current study, for the first time, we evaluated the prevalence of PNPLA3 and TM6SF2 polymorphisms through two different population settings. In other words, by including NAFLD cases from both clinic-based and population-based recruitments, this investigation aimed to answer the question of whether PNPLA3 rs738409 and TM6SF2 rs58542926 are effective on either the initial development of steatohepatitis or its further progression to severe liver diseases. This study has a few limitations. While the sample size for the PB-NAFLD and control was calculated, the study could benefit from a higher number of participants, especially in the CB-NAFLD group. Another limitation is the retrospective data collection in the CB-NAFLD recruitment, which limited the availability and quality of the participants’ clinical data. Finally, the liver disease severity was not assessed in the PB-NAFLD using fibrosis assessment by valid modalities such as liver stiffness measurement.
In conclusion, this study observed the association of PNPLA3 rs738409 with NAFLD in clinic-based recruitment; however, the same was not true for population-based recruitment. Also, TM6SF2 rs58542926 was not found as a modifier of NAFLD and NAFLD-related liver diseases in the Iranian population. More studies with population-based sampling are needed to elucidate the role of genetic variations in the initial development of NAFLD and the progression of liver diseases such as steatohepatitis and liver fibrosis.