1. Background
Hepatitis B virus (HBV) infection remains a significant public health burden worldwide, affecting over 257 million people, and is closely associated with developing liver cirrhosis and hepatocellular carcinoma (1). HBV is a small enveloped DNA virus that establishes its genome in the nucleus of infected hepatocytes by forming the HBV DNA minichromosome, known as the covalently closed circular DNA (cccDNA) (2). The cccDNA serves as a template for the transcription of all viral RNAs, playing a critical role in the viral life cycle and contributing to the persistence of infection and relapse (3). Although effective treatment strategies based on interferon and nucleos(t)ide analogs suppress HBV replication, they do not target cccDNA, challenging the complete eradication of HBV (4). Therefore, the only way to cure chronic HBV infection is to eliminate cccDNA from infected hepatocytes.
MicroRNAs (miRNAs) are a large class of small non-coding RNAs (21 - 25 nucleotides) that regulate gene expression post-transcriptionally by targeting mRNAs (5). Growing evidence suggests that miRNAs may regulate intrahepatic cccDNA expression and play a role in the natural history of chronic hepatitis B (CHB). They can directly affect HBV replication by binding to HBV transcripts or indirectly by targeting cellular factors involved in HBV replication (6). For example, miR-548ah promotes HBV replication and expression by targeting histone deacetylase 4 (HDAC4) (7), which may be related to the deacetylation state of histone binding to cccDNA. Later, Zhang et al. showed that miR-449a directly targeted cAMP-responsive element binding protein 5 (CREB5), which, in turn, induces the expression of farnesoid X receptor α (FXRα). This factor regulates cccDNA activity in specific HBV replication systems (8). While most CHB patients belong to the five phases in the natural history of HBV defined by the EASL in 2017, some individuals do not meet any of the usual immune states and are known to be in the "grey zone (GZ)" (9).
2. Objectives
The objective was to investigate the regulatory role of miRNAs on cccDNA expression in CHB-GZ patients, establish a miRNA-mRNA interaction network, and provide a theoretical basis for finding new therapeutic targets.
3. Methods
3.1. Patients and Liver Tissue Samples
Liver tissues were collected from 13 CHB-GZ patients at the Digestive Diseases Unit of the Virgen del Rocío University Hospital (HUVR). Patients with other liver diseases and viral infections were excluded from the study. The research was approved by the Ethics Committee of the Virgen Macarena-Virgen del Rocío University Hospitals (Approval ID: CEI-HUVM-VR-30/04/2019), and each patient signed an informed consent form. The biopsies were snap-frozen in liquid nitrogen and stored at -80°C until use.
3.2. RNA Extraction
Total or small RNA extractions from tissue samples were performed using the mirVana™ miRNA Isolation Kit (Ambion® Life Technologies, CA, USA) according to the manufacturer's instructions. The total RNA concentration and purity of each specimen were quantified using a NanoDrop™ 2000c spectrophotometer (Thermo Scientific). The purity was estimated by the absorbance ratio at 260 to 280 nm.
3.3. Quantification of Intrahepatic cccDNA
Intrahepatic DNA was isolated from liver tissues using the MasterPure™ DNA and RNA purification kit (Epicenter, Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. The amount of purified DNA was measured using a spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Total intrahepatic cccDNA was determined by real-time PCR, and quantification was performed using the ViiA 7 real-time PCR system mediated by a pair of cccDNA-selective primers and probe. The primers and the probe are shown in Table 1. The quantification range is between 5 × 101 and 5 × 107 copies per reaction for intrahepatic cccDNA. The copies of the viral genome were normalized against the number of cells, estimated by the copy number of the human β-globin gene.
| cccDNA | Sequence | Position |
|---|---|---|
| Forward | CCGTGTGCACTTCGCTTCA | 1577 – 1595 |
| Reverse | GCACAGCTTGGAGGCTTGA | 1884 – 1866 |
| Probe | CATGGAGACCACCGTGAACGCCC | 1609 – 1631 |
3.4. Microarray Profiling of miRNA and mRNA Expression
Transcriptome-wide miRNA and mRNA expression profiles were determined using the GeneChip™ miRNA 4.0 Assay (Affymetrix, ThermoFisher CA, USA) and the Clariom™ S Assay, human, respectively. Sample labeling, microarray hybridization, and washing were performed as recommended by the manufacturer. Finally, the arrays were scanned using the Affymetrix GCS 3000 scanner (Affymetrix, Santa Clara, CA, USA), and signal values were evaluated using the Affymetrix® GeneChip™ Command Console Software (10).
3.5. Screening of Candidate DE-miRNAs and DE-mRNAs
Thirteen CHB-GZ patients' liver tissues were divided into a reference group (n = 7) with cccDNA < 1 copy/cell and a control group (n = 6) with cccDNA ≥ 1 copy/cell, based on the level of cccDNA intrahepatic. We used one copy per cell of cccDNA intrahepatic as the grouping standard because the mean value of cccDNA in HBV-infected patients is 0.01 to 1 copy per cell (11). Moreover, Liang et al. revealed that baseline HBV intrahepatic cccDNA > one copy/cell independently predicts liver inflammation (12). Differentially expressed miRNAs (DE-miRNAs) and mRNAs (DE-mRNAs) between the two groups were selected using the "limma” package in R software. Raw data were initially normalized using the quantile algorithm. The threshold points for DE-miRNAs and DE-mRNAs were |logFC| > log1.5 and P < 0.05.
3.6. Functional Enrichment Analysis
We used the “ClusterProfiler” package in R to perform functional enrichment analysis on the gene ontology (GO) (13) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) (14) pathway to determine the roles played by DE-mRNAs. The GO analysis included three categories: Biological process (BP), cellular component (CC), and molecular function (MF). Statistically significant differences for the enrichment analysis were defined as P < 0.05.
3.7. Construction of miRNA-mRNA Network
We used miRTarBase (15) to investigate the interaction of candidate DE-miRNAs with DE-mRNAs. The miRNA-mRNA network was constructed and analyzed using Cytoscape software (16) (R package). We calculated the Spearman correlation coefficient (r) between miRNA-mRNA target genes. A strong correlation was considered when r < -0.7 and P < 0.01.
3.8. Statistical Analysis
Quantitative data were expressed as mean ± standard deviation. We used SPSS (version 25.0) and R software (version 4.0.5) for statistical analyses. Spearman correlation coefficients were calculated to evaluate the correlations. We considered P < 0.05 to be statistically significant.
4. Results
4.1. Heterogeneity Test and Screening of Candidate DE-miRNAs and DE-mRNAs
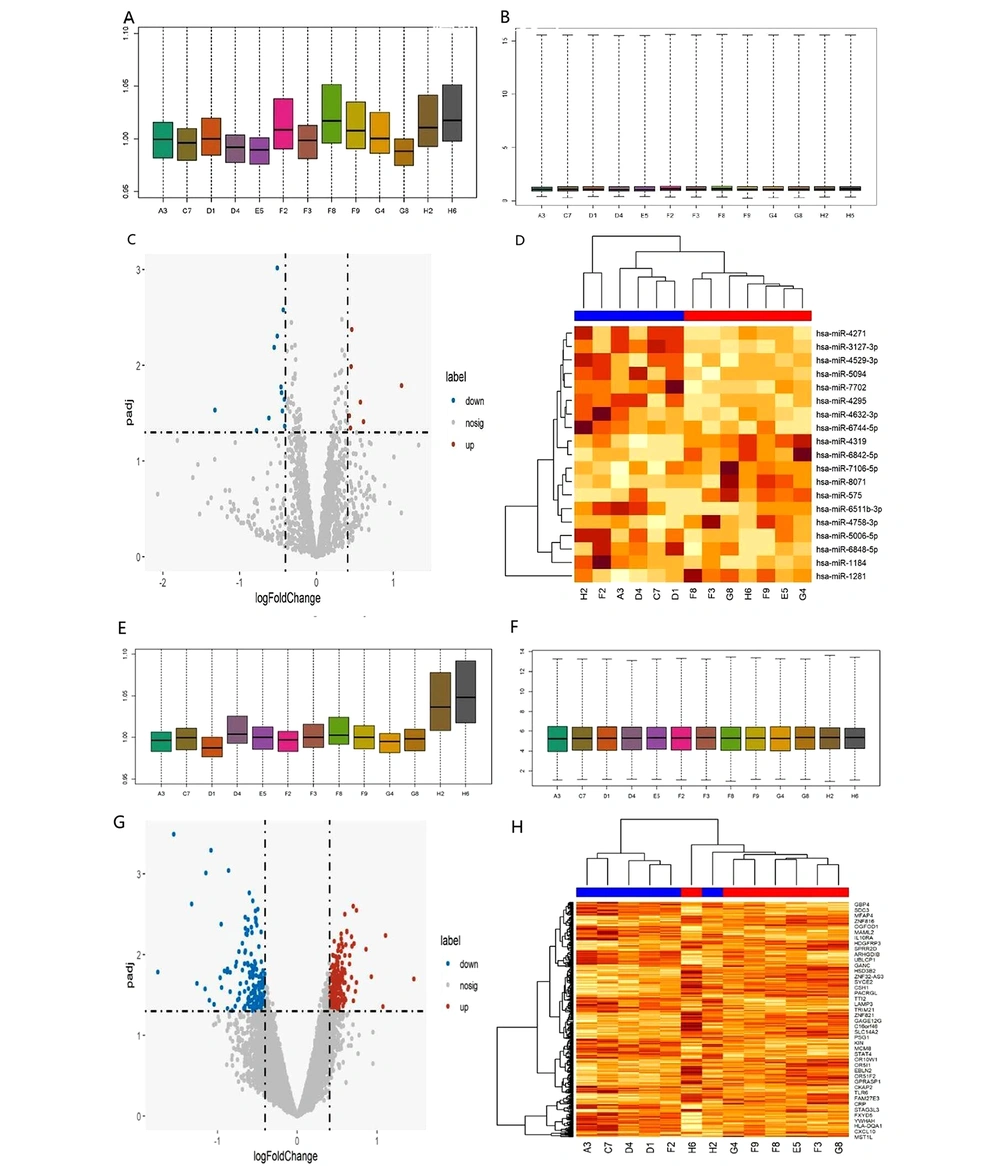
The robust multichip average (RMA) normalization method was used to standardize the data of DE-miRNAs and DE-mRNAs. Then, the “limma" package of the Bioconductor analysis tool was used to make preprocessing and normalized box plots. In comparing CHB-GZ patients with intrahepatic cccDNA < 1 copy/cell and ≥ 1 copy/cell, 19 DE-miRNAs were selected, including 12 down-regulated miRNAs and seven up-regulated. Among them, miR-4295 and miR-4271 were down-regulated, while miR-1281, miR-8071, and miR-575 were up-regulated. A total of 340 DE-mRNAs were obtained, among which 160 were down-regulated and 180 were up-regulated. Among them, ZNF224 and SMYD5 were up-regulated, whereas KPNA2, GPN2, TMEM184B, and POLD3 were down-regulated. The identification of DE-miRNAs and DE-mRNAs is shown in Figure 1.
Identification of DE-miRNAs and DE-mRNAs. A, Box plot of preprocessed miRNA data. B, Box plot of normalized miRNA data. C, Volcano plot of DE-miRNAs. D, Heatmap of DE-miRNAs. E, Box plot of preprocessed mRNA data. F, Box plot of normalized mRNA data. G, Volcano plot of DE-mRNAs. H, Heatmap of DE-mRNAs.
4.2. Functional and Pathway Enrichment Analysis
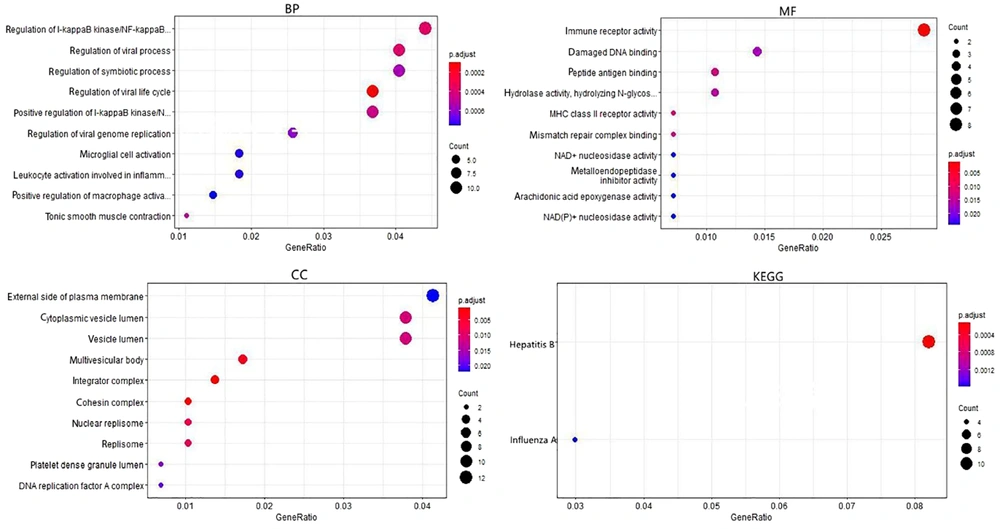
The GO terms were significantly enriched in the biological process of the virus, such as the regulation of the viral life cycle, viral process, viral genome replication, and regulation of the kinase I-kappa B/NF-kappa B. The most enriched MF category in the GO terms was "immune receptor activity," and in the CC category, it was the "outer side of the plasma membrane." KEGG pathway enrichment analysis revealed that hepatitis B and influenza A infections were significantly enriched pathways (Figure 2).
4.3. Construction of miRNA-mRNA Regulatory Network
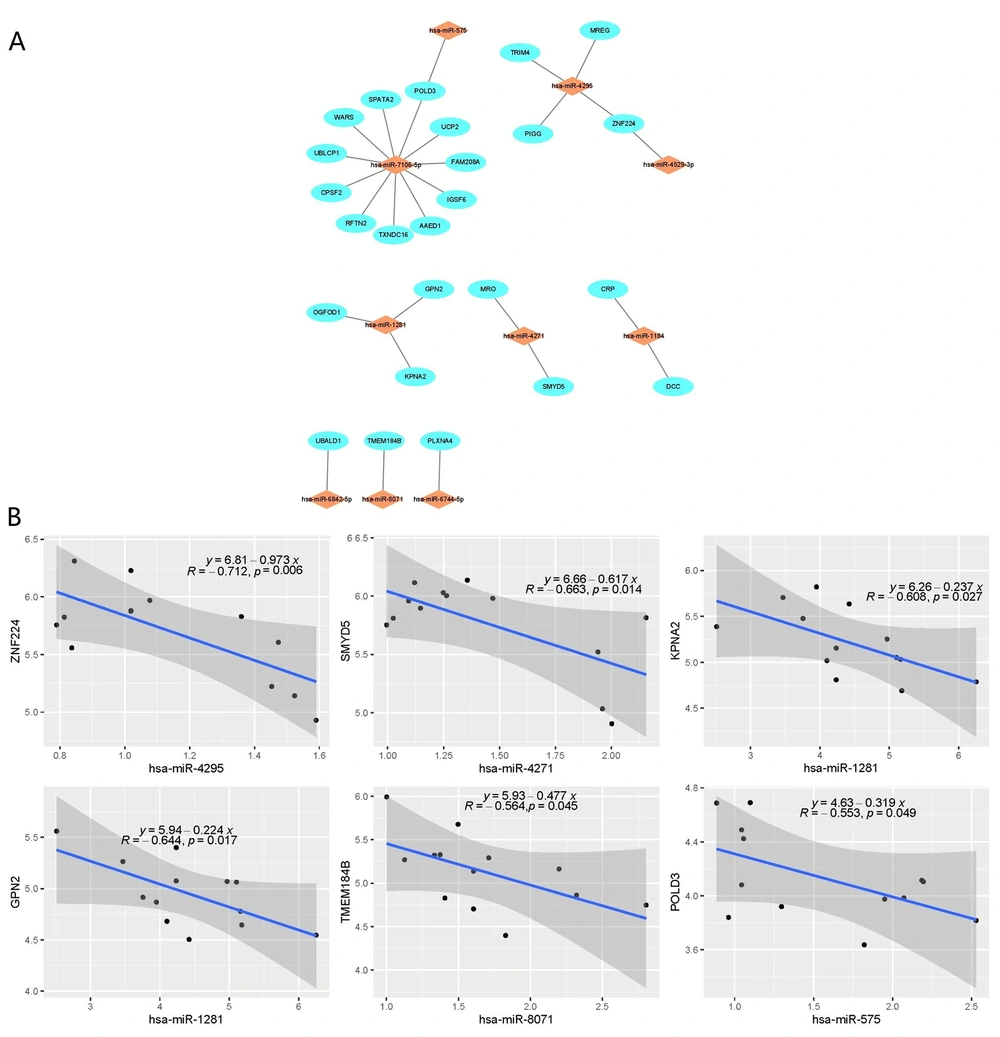
We used the miRTarBase database to evaluate the correlation between the DE-miRNAs and the corresponding target DE-mRNAs. It is well known that target genes are inversely regulated by miRNAs. The higher the correlation coefficient, the greater the mutual regulation effect (17). Based on this theory, we calculated the Pearson correlation coefficient between 19 DE-miRNAs and 340 DE-mRNAs and chose those predicted miRNAs that correlated negatively with target genes (r < 0) and the interaction pairs that exist in miRTarBase for further research (Figure 3A). Subsequently, we selected six pairs of miRNA-mRNA interactions (miR-4295 - ZNF224, miR-4271 - SMYD5, miR-1281 - KPNA2, miR-1281 - GPN2, miR-8071 - TMEM184B, and miR-575 - POLD3) with P < 0.05. The correlation of the above six significant interaction pairs is shown in Figure 3B. Finally, a significantly strong negative correlation was found between the miR-4295-ZNF224 interaction pair with a cut-off value of r < -0.7 and P < 0.01.
5. Discussion
The miRNA-mRNA regulatory network is associated with the initiation and progression of various human diseases (17-19). However, the miRNA-mRNA regulatory network that regulates intrahepatic cccDNA of CHB-GZ patients is still unknown. In this work, we aimed to identify several miRNAs, their target genes, and miRNA-mRNA regulatory network that may be involved in regulating the expression of cccDNA in patients with CHB-GZ through a series of bioinformatic analyses. These analyses provide us with a theoretical basis for locating new therapeutic targets.
A total of 19 DE-miRNAs, including 12 down-regulated miRNAs and seven up-regulated miRNAs, and 340 DE-mRNAs, including 160 down-regulated mRNAs and 180 up-regulated mRNAs, were screened. A series of bioinformatic analyses established a possible miRNA-mRNA regulatory network. A total of six pairs of miRNA-mRNA (miR-4295 - ZNF224, miR-4271 - SMYD5, miR-1281 - KPNA2, miR-1281 - GPN2, miR-8071 - TMEM184B, and miR-575 - POLD3) were obtained, among which miR-4295 - ZNF224 showed a relatively high correlation.
ZNF224 is a Kruppel-associated box zinc finger protein (KRAB-ZFP) family member, the largest family of transcriptional regulators in higher vertebrates. It is characterized by an N-terminal KRAB domain and a C-terminal array of DNA-binding zinc fingers (20). Despite their numerical abundance, the functions of KRAB-ZFP have not been clearly defined for a long time. However, cumulative data indicate that some processes involve imprinting, cell differentiation, metabolic control, and gender dimorphism (20). The ZNF224 gene is differentially expressed in human tissues and cells. According to the distinctive cellular environments, it exhibits different essential functions in transcription or post-transcriptional gene regulation (21). ZNF224 was initially identified as a transcriptional repressor involved in the specific silencing of genes through chromatin-modifying activities, the recruitment of the corepressor KRAB-associated protein 1 (KAP1), and the protein arginine methyltransferase 5 (PRMT5) in the promoter of its target genes (21). In carcinogenesis, ZNF224 was assigned a dual role of tumor suppressor and oncogene, thus causing apoptosis or stimulating cell proliferation (22). Its function as an oncogene was discovered in bladder cancer, breast cancer, and chronic lymphocytic leukemia (22). However, ZNF224 in chronic myeloid leukemia is a tumor suppressor that can promote apoptotic gene expression and suppress anti-apoptotic gene expression to improve drug resistance (23).
MiR-4295 is a newly discovered miRNA located on chromosome 10q25.2. It is a unique miRNA with many biological functions (24). MiR-4295 is widely expressed in vivo, participating in the biological processes of multiple cancers. At the same time, it promotes cancer by affecting seven cellular activities, including cell proliferation, cell migration, cell cycle, cell invasion, epithelial-mesenchymal transition (EMT), and apoptosis (24). For example, miR-4295 promotes the growth of bladder cancer cells by targeting the B-cell translocation gene 1 (BTG1) (25). It contributes to cell invasion and proliferation of pancreatic ductal adenocarcinoma through glypican-5 (GPC5) (26). It can also promote cell migration, proliferation, and invasion of osteosarcoma by targeting interferon regulatory factor 1 (IRF1) (27). Although miR-4295 is dysregulated in various cancers, it has also been found to inhibit cancer (24). The expression of miR-4295 is related to prognosis and can be affected by numerous factors connected to the therapeutic effects of various drugs (24). The target genes of miR-4295 include TP63, USP28, RUNX3, GPC5, CDKN1A, BTG1, NPTX1, LRIG1, and PTPN14 (25, 28, 29). In addition, downstream genes of miR-4295 also include SOS2, EREG, VEGF, VEGF-A, FLT1, FLT2, and FOXF1 (30, 31). Three signaling pathways related to miR-4295, including N-myc/miR-4295/RUNX3, miR-4295/GPC5/Wnt/β-catenin, and miR-4295/EGFR/PI3K/Akt, have been identified (24). The Wnt/β-catenin pathways are well-known in hepatitis B infection and hepatocellular carcinoma.
Although no research has revealed the relationship between miR-4295 and ZNF224 to the HBV infection process, the enrichment function of the GO and KEGG pathway is primarily related to the biological process of the hepatitis B virus. More enriched GO terms were related to regulating the viral life cycle, the viral process, and the viral genome replication. Enrichment analysis of the KEGG pathway suggested that these predicted targets were related to hepatitis B and influenza A. According to an analysis, annual influenza vaccination reduces the risk of hospitalization and death in CHB patients, suggesting possible impacts of influenza-targeted therapy on HBV infection (32).
5.1. Limitations
While we conducted a thorough analysis and identified several potential miRNA-mRNA axes that may regulate intrahepatic cccDNA in CHB-GZ patients, we acknowledge some limitations to this study. Firstly, the sample size was relatively small, consisting of only 13 liver tissue samples. Additionally, the miRNA-mRNA regulatory axis we identified has yet to be validated by in vivo or in vitro experiments. Therefore, larger clinical samples and corresponding experiments are required to confirm our results in the future. Long-term follow-up is also necessary to observe disease progression.
5.2. Conclusions
In summary, we have identified a potential miRNA-mRNA regulatory axis related to intrahepatic cccDNA in patients with CHB-GZ. Our study suggests that decreased expression of miR-4295 in CHB-GZ patients with a low level of cccDNA is associated with increased expression of ZNF224, which is involved in the biological process of HBV replication and the regulation of its life cycle. These findings could provide new potential targets for the treatment of CHB infection. However, larger clinical samples and further experiments are needed to verify our findings.