1. Background
Liver cancer is known as one of the most malignant cancers, with 905000 new cases and 830000 deaths in 2020 worldwide, making it the seventh most common and the second most lethal cancer globally. Hepatocellular carcinoma (HCC), comprising 75 - 85% of all liver cancer cases, is the dominant form of liver cancer (1). The major risk factors of HCC include hepatitis B virus, hepatitis C virus, diabetes, obesity, and alcohol, making HCC more common in less developed regions of the world (1, 2).
Liver tumor cells possess very efficient resistance mechanisms against drugs; therefore, chemotherapy, as the most applied treatment for cancer, has limited usage against HCC (3). Consequently, three types of surgery, including liver transplantation, liver resection, and liver ablation, comprise the most effective treatments for HCC, none of which offers a permanent treatment, and each possesses its own limitations (4). Despite the limited usage of chemotherapy, certain drugs, such as etoposide (ETO), are found to be influential against HCC (3, 5). Etoposide, a plant-based anticancer agent, halts cancer cell growth and causes cell death through binding to deoxyribonucleic acid (DNA) and to topoisomerases II enzyme resulting in preventing DNA double-strand reconnection (6, 7). As ETO brings about several side effects on patients, seeking novel methods, such as combinational therapy, is considered a way to reduce its adverse effects while simultaneously enhancing its antitumor impacts (8).
As a tumor suppressor, p53 plays important roles in cell cycle regulation, DNA repair, and apoptosis induction; however, the damaged DNA is not fixed (9, 10). The p53 mutation is observed in about 50 - 60% of all cancers, including HCC, which are either gain of function or loss of function mutations, both of which could lead to tumorigenesis (11). In addition, it is observed that the p53 gene mutations enhance the chemo-resistance of liver cancer cells (12). Therefore, it is necessary to monitor the p53 status when studying cancer.
Plant-derived compounds, such as polyphenols, attract much attention regarding their ability to ameliorate diseases, such as cardiovascular diseases (CVDs), neurodegenerative diseases, and cancers (13-15). Quercetin (Que) is the most abundant flavonoid from the large polyphenol family that has shown anticancer effects. Regarding recent studies, Que induces apoptosis and inhibits the proliferation and migration of different cancer cell lines (16). The main pathways affected by Que are extracellular signal-regulated kinase 1/2 (Erk1/2), phosphatidylinositol 3-kinase (PI3K), and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB) pathways all of which have significant roles in the expression of numerous cancer-related genes (17-19). It is believed that Que performs its anticancer effects through its antioxidant and anti-inflammatory activities (20, 21). Although Que has been frequently studied, the effects of Que in combination with ETO on the HCC cell line are yet to be inspected.
2. Objectives
This study aimed to investigate the effects of the combination of Que and ETO on apoptosis induction and on the expression of p53 protein in the HCC cell line.
3. Methods
3.1. Cell Culture and Reagents
The HCC HepG2 cell line, purchased from Pasteur Institute (Tehran, Iran), grows properly in Dulbecco’s modified eagle’s medium (DMEM) culture medium, containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin antibiotics. However, one day before the assay started, it was replaced with a fresh medium, excluding FBS. Cell incubation was performed at 37°C and 5% CO2. Que powder was bought from Sigma Aldrich (United States) and was solved into dimethyl sulfoxide (DMSO) immediately before treatment. The concentration of DMSO in the cell culture medium was constantly maintained below 1% (V/V) to avoid any adverse effects on cell viability. To obtain the desired concentration, the Que solution was diluted in DMEM.
3.2. Cell Viability (MTT) Assay
In this study, 104 HepG2 cells were seeded into 96-well plates and were incubated overnight (37°C). Over the following day, the culture medium for both cell lines was altered with a medium containing Que (25, 50, and 100 uM). The medium was then eliminated, and the medium containing MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) solution was added to the cells for 4 hours. Afterward, the MTT-containing medium was changed with 150 mL DMSO added to each well seeded with cells and shaken until the formazan crystals were solved entirely. The optical density for each well was read in 570 nM. The cells were treated with Que for time courses of 24, 48, and 72 hours for this experiment. The results were then converted to the %viability of cells.
3.3. Relative Gene Expression Determination (Quantitative Reverse Transcription Polymerase Chain Reaction)
Total ribonucleic acid (RNA) isolation was performed from 1 million cells cultured in 6-well plates using a Yekta Tajhiz Azma RNA extraction kit (Iran). The concentration and the purity of the final isolate were determined using Nanodrop 2000 instrument (Thermo Fisher Scientific, Wilmington, DE, United States), and its integrity was examined through 1.5% agarose gel electrophoresis. The RNA isolates were then utilized for complementary DNA (cDNA) synthesis, using Yekta Tajhiz Azma cDNA synthesis kit, with the final reaction volume of 20 μL. The final cDNA products were stored at -70°C. The relative expression of B-cell lymphoma 2 (Bcl-2) associated X-protein (Bax) and Bcl-2 genes were assessed through quantitative reverse transcription polymerase chain reaction (PCR), utilizing Amplicon SYBR green kit (Thermo Fisher Scientific, USA), with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the internal reference gene and the final volume of 20 μL. The QuantStudio 3 Applied Biosystems (Massachusetts, United States) PCR instrument performed the reaction steps. The sequences of specific primer pairs, newly designed for this study, are as follows:
Bax (F: 5′-CAGGGGCCCTTTTGCTTCA-3′, R: 5′-ACGGCGGCAATCATCCTCT-3′).
Bcl-2 (F: 5′-GGATAACGGAGGCTGGGATG-3′, R: 5′-TGACTTCACTTGTGGCCCAG-3′).
GAPDH (F: 5′-ACCCTTAAGAGGGATGCTGC-3′, R: 5′-CCCAATACGGCCAAATCCGT-3′).
3.4. Western Blot Analysis
Equal amounts of protein were collected from cell extracts previously homogenized with RIPA buffer (containing HEPES, NaCl, EDTA, and Triton X100) and separated through sodium dodecyl-sulfate-polyacrylamide gel electrophoresis. The proteins were then transferred to a polyvinylidene difluoride membrane and closed by skim milk-containing tris-buffered saline with 0.1% Tween® 20 detergent for an hour (25°C). Utilizing specific primary antibodies against Bax, Bcl-2, and p53 (all obtained from Cell Signaling, Danvers, Massachusetts, United States) with incubation at 4°C overnight and secondary antibodies at room temperature for an hour, western blotting was conducted. The bands were visualized using an enhanced chemiluminescence system and finally quantified using Image J software (version 1.52). The normalization of protein levels was performed with β-actin as the control.
3.5. Flow Cytometry
Annexin V-FITC/PI kit (IQ Products, Groningen, Netherlands) was utilized to carry out apoptosis assay. Briefly, the cells were centrifuged, and the cell pellet was suspended in calcium buffer with Annexin V-FITC and incubated at 4°C for 20 minutes. Afterward, the previous buffer was discarded, the same buffer containing PI was added to the cells, and incubation was performed at 4°C for 10 minutes. The samples and the subsequent data were analyzed using a flow cytometer (Becton, Dickinson, San Jose, CA, United States) and FlowJo software (version 10).
3.6. Caspase-3 and Caspase-9 Activities
The activities of caspase-3 and caspase-9 were measured through the incubation of the lysates of Que-treated cells with colorimetric substrates for an hour and, finally, the evaluation of the absorbance, caused by the cleavage of the substrate through caspases activities, at 405 nm.
4. Results
4.1. Indication of Synergy by Que and ETO on the Inhibition of HepG2 Cell Growth
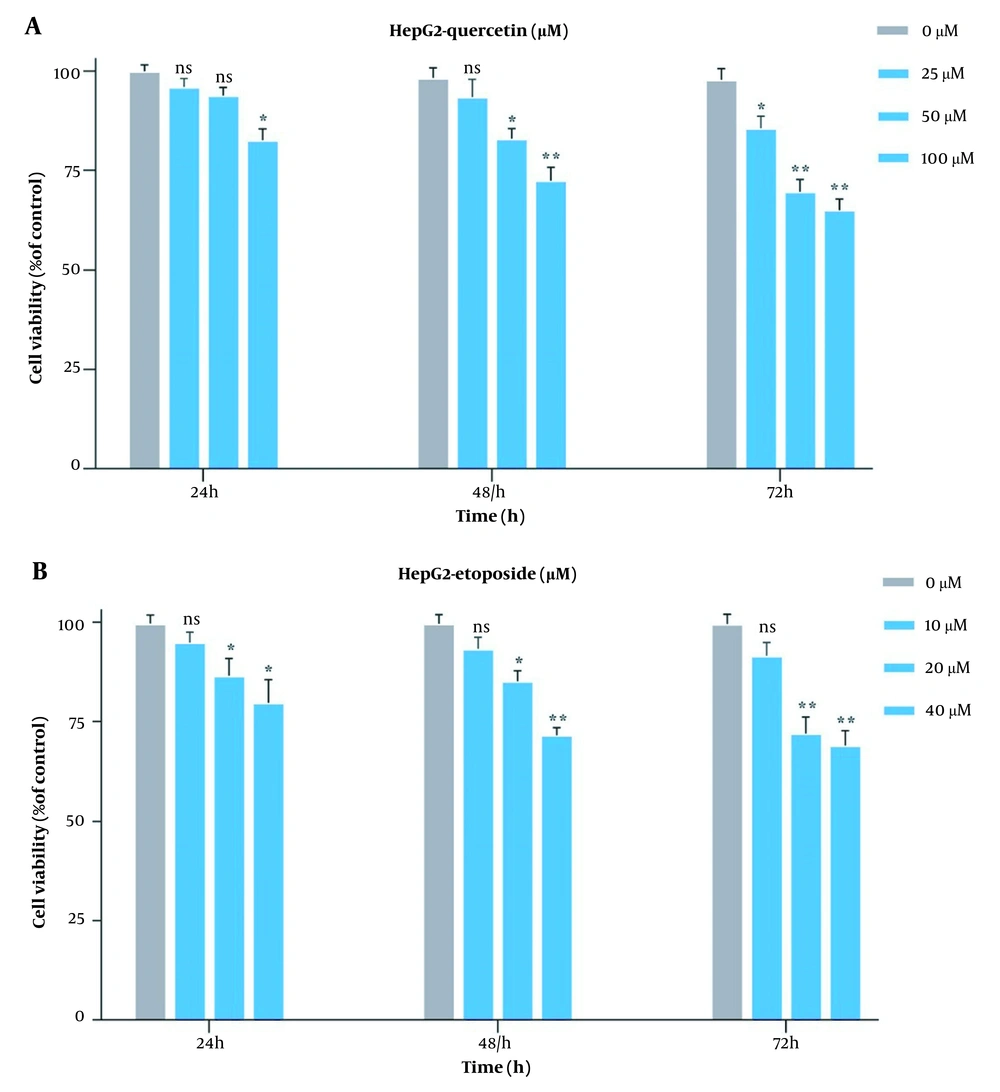
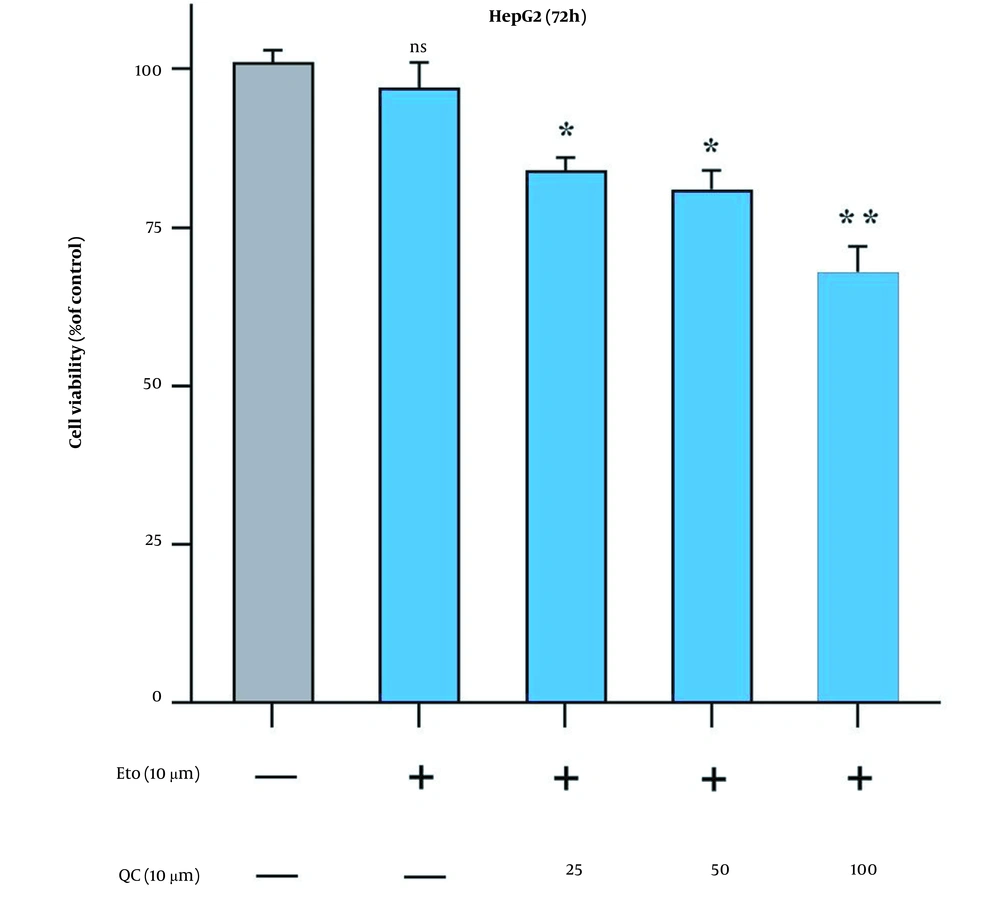
Originally, to obtain the optimal concentration for each drug, this study examined the HepG2 cell growth when treated with Que and ETO individually through the MTT assay. The initial concentrations of ETO were 25, 50, and 100 µM. The initial concentrations of Que were 10, 20, and 40 µM. Concerning the results, both Que and ETO showed dose-dependent and time-dependent behaviors on cell growth. The current study’s results showed that in 24 hours, the only effective dose for Que was 100 µM; nevertheless, the 50 µM dose was appended in 48 hours, and all doses showed significant effects on cancer cell growth in 72 hours. Moreover, 100 µM was the concentration of Que with the highest inhibitory effects on cancer cells (32% growth inhibition in 72 hours) (Figure 1A). The 10 µM concentration of ETO showed no significant effects on cell growth in either of the time courses; however, the 20 and 40 µM doses both significantly reduced the cancer cell growth in all time periods (Figure 1B). It is worth mentioning that no concentration of Que and ETO reached 50% growth inhibition (i.e., the half maximal inhibitory concentration). The 10 µM concentration of ETO was selected for combination with 25, 50, and 100 µM of Que. All combinations decreased cancer cell growth higher than ETO alone, with the combination of Que 100 µM and ETO 10 µM presenting the highest effect (Figure 2). According to combination indices, Que showed synergy with ETO through all present concentrations. However, the combination showing the lowest combination index (0.48), comprising the most and least effective concentrations of Que (100 µM) and ETO (10 µM), respectively, was selected for the rest of the experiments (Table 1).
Evaluation of impacts of quercetin and etoposide on the growth of HepG2C through MTT assay; A, treatment of HepG2 cells with three bottom-up concentrations of quercetin for 24, 48, and 72 hours; B, treatment of HepG2 cells with three bottom-up concentrations of etoposide for 24, 48, and 72 hours (ns: Not significant; * P < 0.05, ** P < 0.01).
Comparison of effects of quercetin and etoposide to etoposide alone on the growth of HepG2 cells through MTT assay; treatment of cancer cells with 10 µM of etoposide combined with 25, 50, and 100 µM of quercetin for 72 hours (ns: Not significant; * P < 0.05, ** P < 0.01 compared to control).
| Quercetin (25 μm) | Quercetin (50 μm) | Quercetin (100 μm) | |
|---|---|---|---|
| Etoposide (10 μm) | 0.93 | 0.76 | 0.48 |
a HepG2 cells were incubated with 10 μM of etoposide and various concentrations of quercetin. The combination index (CI) was calculated by CompuSyn software (version1.0.1). CI < 1.0, CI = 1.0, and CI > 1.0 represent synergism, an additive, and antagonism, respectively.
4.2. Potentiation of Effect of ETO by Que on HepG2 Cell Apoptosis Induction
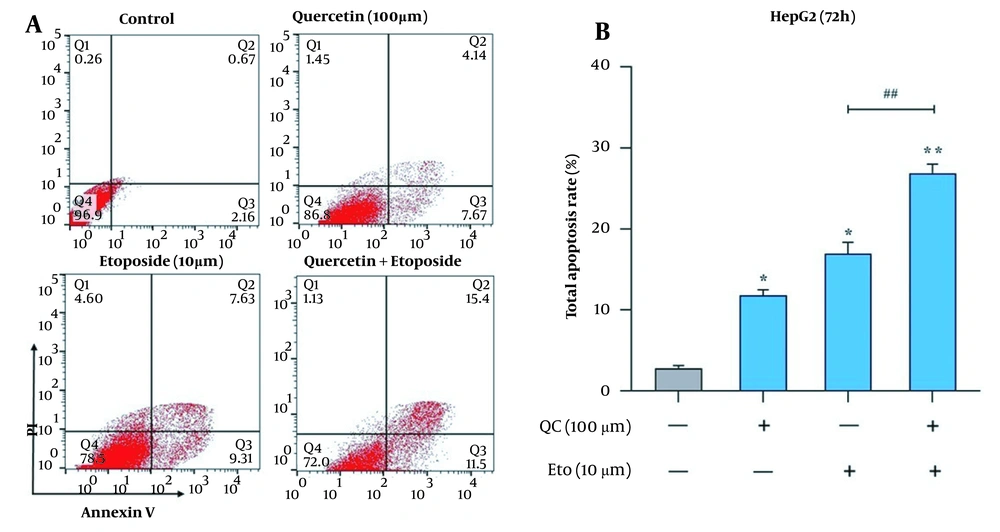
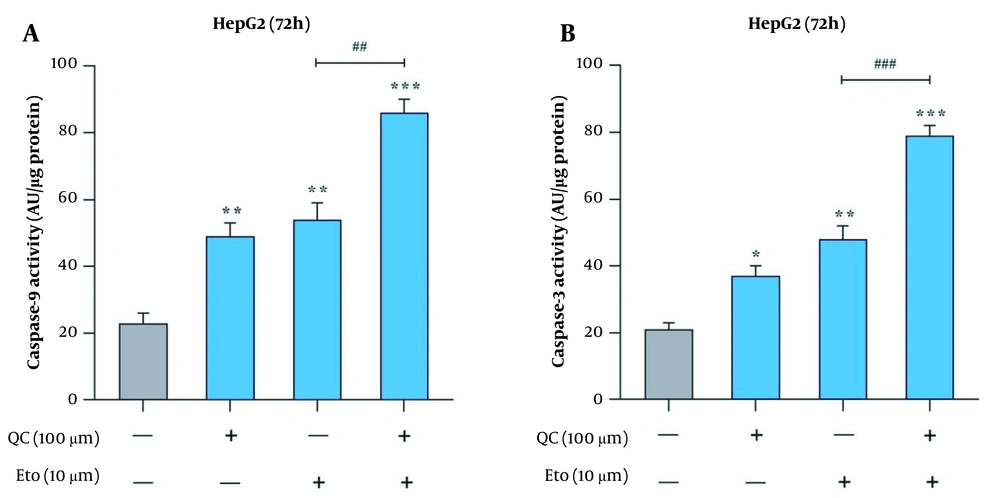
The effects of Que and ETO on the apoptosis rates of HepG2 cells were examined through two tests, flow cytometry and caspase-9 and caspase-3 activity assay. Through Annexin/PI double staining, it was observed that both Que and ETO could increase the programmed death rates of the investigated cells individually, with Que raising it to 11.8% and ETO to 16.9%. Furthermore, after treatment with the Que + ETO combination, the rate of programmed cell death reached 26.9%, showing synergy between Que and ETO activities. Figure 3A and B illustrates the obtained results in this regard. Regarding caspase-9 activity, the individual concentrations of Que and ETO showed 49% and 54% activity, respectively; nevertheless, with the Que + ETO combination, the caspase-9 activity exceeded 86% (Figure 4A). In addition, both Que and ETO alone enhanced the activity of caspase-3 to 37% and 48%, respectively. As observed in Figure 4B, through exposure to the Que + ETO combination, the caspase-3 activity underwent a further enhancement and reached 79%.
Effects of quercetin and etoposide on the apoptosis of HepG2 cells through flow cytometry; A, flow cytometry plot of the effects of quercetin and etoposide individually and in combination on the HepG2 cell death; B, columnar plot of flow cytometry for quercetin and etoposide (* P < 0.05, ** P < 0.01 compared to control; ## P < 0.01 compared to etoposide alone).
Activities of caspases in HepG2 cells treated with quercetin and etoposide; A, caspase-9 activity in HepG2 cells after treatment with quercetin and etoposide alone and in combination; B, caspase-3 activity in HepG2 cells after treatment with quercetin and etoposide alone and in combination (* P < 0.05, ** P < 0.01 compared to control; ## P < 0.01, ### P < 0.001 compared to etoposide alone).
4.3. Increase in Bax and Reduction of Bcl-2 Gene Expression and Protein Levels by ETO + Que Combination
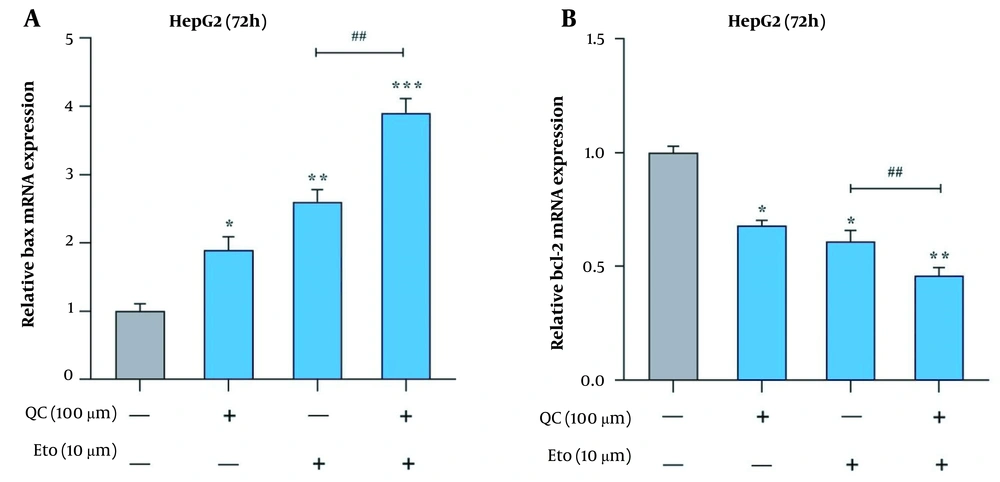
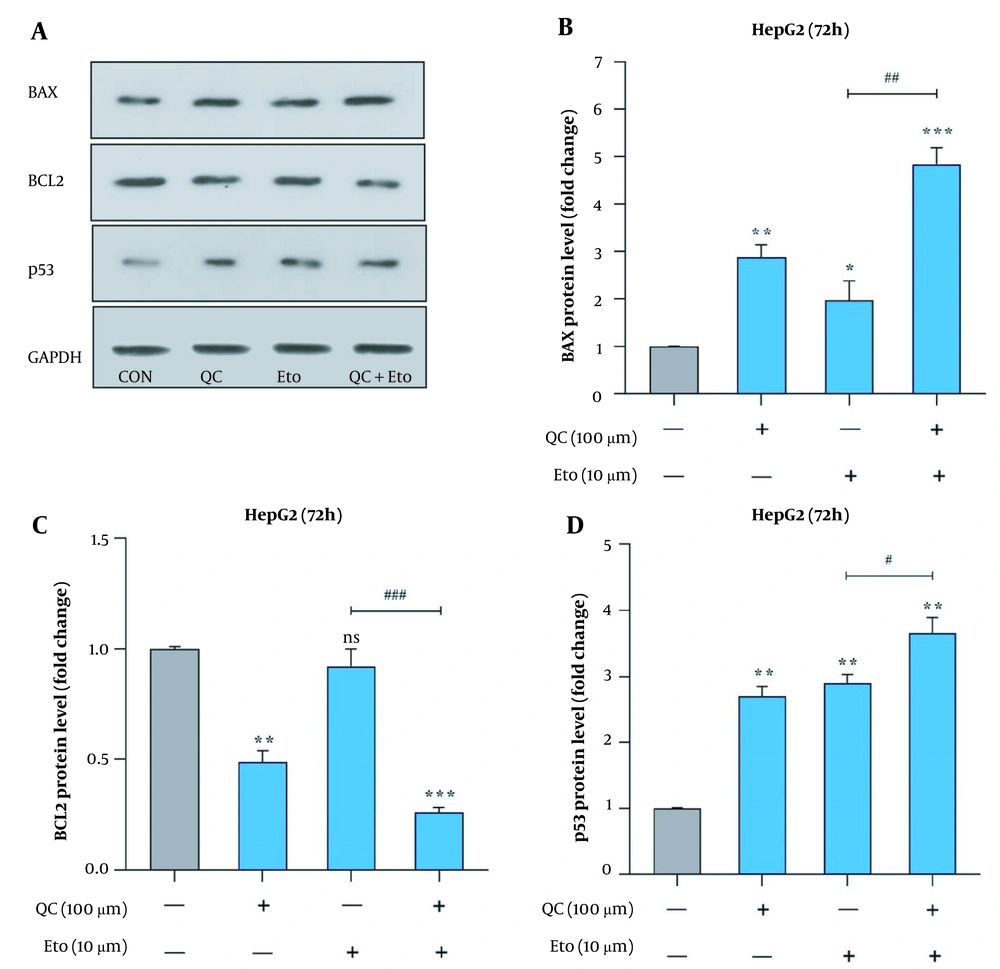
Through real-time PCR, it was observed that the Que + ETO combination adjusted the expression of Bcl-2 and Bax genes toward the promotion of apoptotic cell death. The present study’s results showed that both Que and ETO individually increased the gene expression of Bax to 1.89 and 2.6 folds, respectively; however, the combination showed a further effect on the Bax gene, increasing its expression to 3.9 folds (Figure 5A). Conversely, Bcl-2 expression declined to 0.68 and 0.61 folds after treatment with Que and ETO, respectively; nonetheless, it was lowered to 0.46 folds when exposed to the Que + ETO combination (Figure 5B). The levels of Bax protein were elevated to 2.85, 1.98, and 4.81 folds, with the treatment of Que, ETO, and the ETO + Que combination, respectively. Additionally, it was observed that the rise in Bax protein levels caused by the ETO + Que combination was significantly higher than the ETO treatment alone (Figure 6A and B). Que decreased the protein levels of Bcl-2 to 0.48 folds; nevertheless, ETO demonstrated no significant effects on this protein at all. Furthermore, the ETO + Que combination reduced the Bcl-2 protein expression to 0.26 folds, showing a significant reduction, compared to the untreated control and ETO-treated groups (Figure 6A and C).
Effects of quercetin and etoposide on the relative expression of apoptosis-related genes in HepG2 cells; A, Bcl-2 associated X-protein (Bax) relative gene expression in HepG2 cells treated with quercetin and etoposide alone and in combination; B, B-cell lymphoma 2 (Bcl-2) relative gene expression in HepG2 cells treated with quercetin and etoposide alone and in combination; (* P < 0.05, ** P < 0.01, *** P < 0.001 compared to control; ## P < 0.01 compared to etoposide alone).
BAX, BCL2, and p53 protein levels in HepG2 cells treated with quercetin and etoposide; A, western blot gel image of the effects of quercetin and etoposide on BAX, BCL2, and p53 protein; B, effects of quercetin and etoposide on the BAX protein expression; C, effects of quercetin and etoposide on the BCL2 protein expression; D, effects of quercetin and etoposide on the p53 protein expression (* P < 0.05, ** P < 0.01, *** P < 0.001 compared to control; # P < 0.05, ## P < 0.01, ### P < 0.001 compared to etoposide alone).
4.4. Synergistic Increase of p53 Protein Levels by ETO + Que Combination
This study further explored the effects of Que and ETO on the apoptosis induction of HepG2 cells by evaluating the p53 protein levels. The obtained results showed that exposure to both Que and ETO alone leads to a significant rise in the tumor suppressor p53 protein level in the studied cells. Specifically, in a close manner, Que and ETO promoted the p53 levels to 2.7 and 2.9 folds, respectively. Finally, as apparent in the current study’s results, Que potentiated the effect of ETO on the protein expression of p53, causing a rise of 3.65 folds in the tumor suppressor’s protein levels. Figure 6A and D depicts the results of p53 protein expression.
5. Discussion
Cancer therapy has been a constant challenge for humanity, especially for cancer researchers. There are virtually as many cancers as organs in the human body, any of which has its own characteristics and behavior and its unique treatment (1). The HCC, the most common type of liver cancer, is known for its progressiveness, lethality, and resistance to common cancer therapies (1, 22, 23). Chemo-resistant and radiation-resistant cancers, through specific procedures, develop resistance against different drugs, minimizing the impacts of the therapy (24). In these cases, the implemented drug doses gradually fail, and higher doses are required, leading to extra side effects (25). Therefore, it is logical to find supplementary therapies with minimum adverse side effects and, at the same time, capabilities against cancer (26). Among numerous plant-based compounds, polyphenols attract greater attention by affecting the treatment of CVDs, Alzheimer’s disease, and cancers (27-29).
As a programmed and modifiable cell function, apoptosis plays an important role in the life cycle of a single cell and the wellness of the whole organism (30). Numerous prominent proteins, such as p53, c-Myc, tumor necrosis factor (TNF) family, Fas, Bcl-2 family, mitogen-activated protein kinase family, NF-kB, and caspase family, function inside or outside a cell, playing significant roles in the prevention, induction, or regulation of apoptotic cell death, showing the importance of this cancer-preventive cellular function (31, 32). Bax and Bcl-2 are two important members of the Bcl-2 family, where the former plays an anti-apoptotic role, and the latter is pro-apoptotic (33-35). The diversity of apoptosis-related genes leads to the countless ways of apoptosis malfunction, such as mutations or hyperactivity of anti-apoptotic factors and deficiency in pro-apoptotic factors, through which cancer cells develop mechanisms to excess their survival limit or obtain the ability to recurrently proliferate, even upon a defective DNA (36-39).
Que, a flavonol member of the polyphenol substances family, plays roles in cellular functions, including anti-inflammatory, antioxidant, pro-apoptotic, and, generally, anticancer activities (40-42). Quercetin’s anti-proliferation and pro-apoptotic abilities have been studied in different cancers, presenting its relationship with various cellular apoptosis-related factors, such as Erk1/2 pathways, NF-kB, and TNF-α (43, 44). Furthermore, it has been shown that Que shows its effects by enhancing the antitumor effects of common cancer drugs, such as 5-fluorouracil and cisplatin (45, 46). Compounds possessing such abilities produce major benefits since they help minimize the side effects of chemotherapeutics, maximize the suppressive effects of the chemo-drug on tumor cells, and efface potential chemo-resistance in cancer cells (47, 48). Therefore, the present study investigated whether Que shows any synergy with the widely used chemotherapeutic ETO on the apoptosis induction and reduction of HepG2 cell survival. According to the MTT assay, the effects of both Que and ETO on the cell viability of HepG2 are time- and dose-dependent, where in 72 hours, even the lowest concentration of Que affected the cancer cell viability significantly, suggesting a rather high half-time for Que in cancer cells. Additionally, the present study’s results, similar to a previous study, showed that Que could enhance the anti-proliferative activities of ETO on the HepG2 cancer cells. Different mechanisms have been referred to as the apoptosis-inducing ways of Que.
Catanzaro et al. and Suh et al. both suggested that Que induces cancer cell apoptosis through cell cycle arrest in G2/M and G0/G1 phases, respectively (49-51). Furthermore, the results of the studies by Shen et al. and Li et al. have shown that Que induces the mitochondrial pathway of apoptosis by altering the PI3K/Akt and NF-kB pathways alone and in combination with chemotherapeutics (52, 53). The current investigation explored the rates of apoptotic cell death under treatment with Que and ETO alone and in combination and its potential relationship with the tumor suppressor p53. It was observed that not only Que promoted cancer cell apoptosis alone, but also it increased the effects of ETO on the apoptosis of cancer cells in a synergistic manner. The results of the apoptosis assay were reflected in the caspase-3 and -9 activity assays and in the Bax and Bcl-2 genes’ expression, where the significant rise in both caspases’ activities, the Bax gene expression, and the down-regulation of the anti-apoptotic Bcl-2 gene altogether pointed at ETO and Que as effective drugs for the induction of HepG2 cell apoptosis.
The apoptotic cell death of ETO + Que-treated cells experienced a rise of 24.07%, compared to the control group, proving that the activities of ETO on HepG2 cells were significantly enhanced when combined with Que. However, with the assessment of Bax and Bcl-2 gene expression, it is not believed that it showed any effects of the two factors on cancer cell apoptosis, given the diverse events that might occur between gene expression and protein translation, particularly in an apoptotic cell (54). Therefore, the current study assessed the expression of Bax and Bcl-2 proteins to evaluate the possibility of any connection between these factors and the cancer cell apoptosis induced by the drugs.
The present study showed that the treatment with the combination of ETO + Que led to an elevation in Bax protein level while reducing the expression of Bcl-2 protein, both of which were statistically significant. With the positive relation between the apoptosis of cancer cells and the expression of Bax protein, it is apprehended that the effects of the ETO + Que combination could be exerted through the fluctuations of Bcl-2 family members. However, further studies are needed to investigate the exact mechanisms.
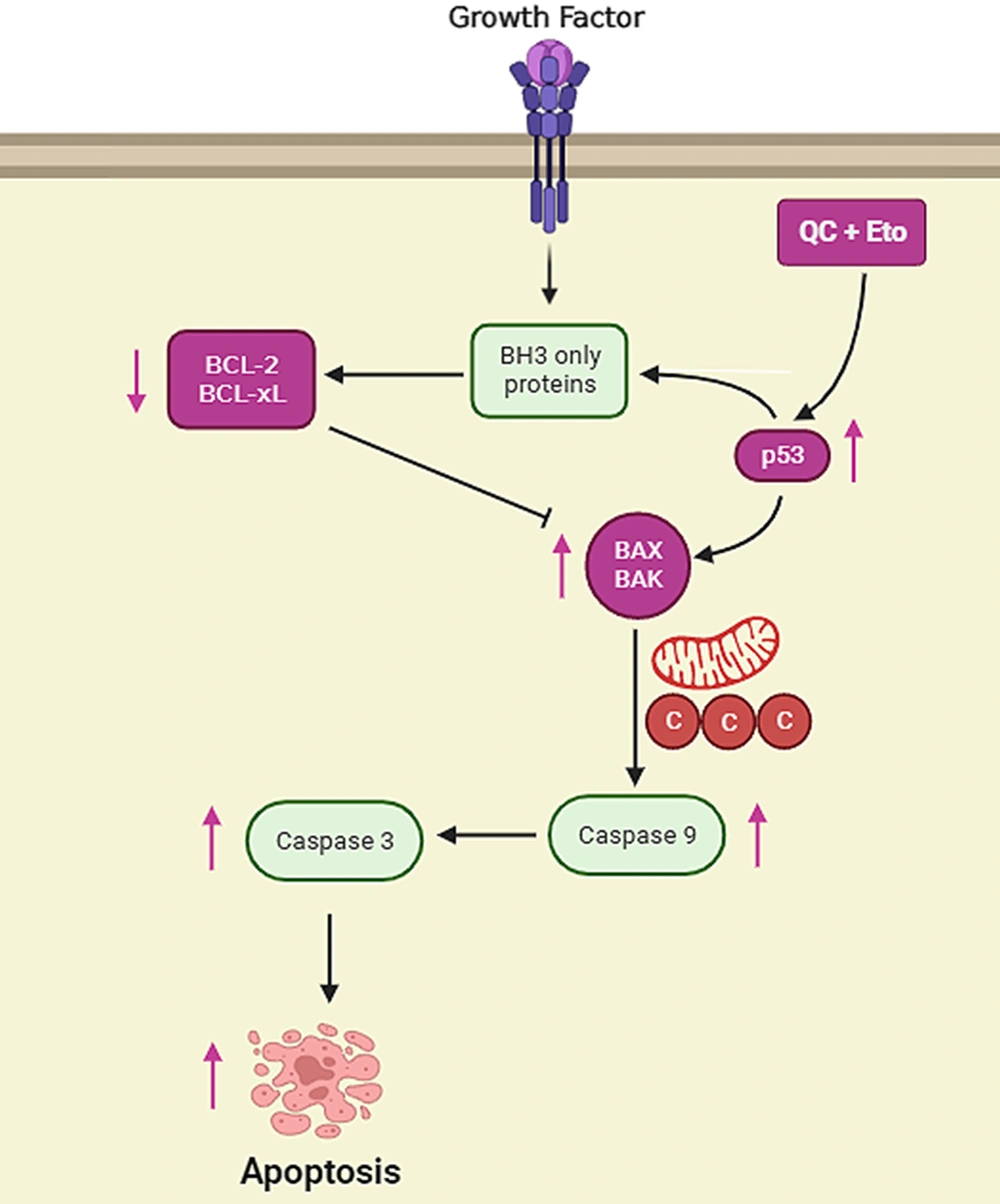
The role of p53 has been investigated in numerous cancer studies, where it is clarified that p53 is a major inducer of apoptosis in defected cells. It has been shown that p53 possesses the ability to induce DNA repair, initiate cell cycle arrest, promote cellular senescence, and even induce cell apoptosis by multiple important targets, such as DNA damage response (DDR), p53 upregulated modulator of apoptosis (PUMA), and TP53-inducible glycolysis and apoptosis regulator (TIGAR), regulating the activities of major cellular factors and processes, such as Bcl-2, Bax, cyclin-dependent kinases, and generally, cell apoptosis (9, 55). Many different defects in cellular processes could lead to p53 activation; however, the high percentage of p53 mutations in different types of cancer prevents it from performing its proper roles in cancerous cells, leaving the cell vulnerable to multiple defects in DNA and other critical parts, without any proper repair (56-58). In the current study, it was shown that p53 protein levels were significantly elevated after treatment with both Que and ETO for 72 hours. In HepG2 cells treated with the Que + ETO combination, p53 protein expression underwent even higher promotion, becoming 0.95 and 0.75 folds higher than Que-treated and ETO-treated cells, respectively. The aforementioned results suggest that the higher apoptosis rates in Que + ETO-treated cells could have occurred through p53 resurrection, leading to higher Bcl-2 gene expression and more caspase activities, driving the cancer cells toward a programmed cell death (Figure 7).
Schematic presentation of apoptosis pathways in HepG2 cells treated with quercetin and etoposide quercetin and etoposide induce apoptosis in HepG2 cells by promoting p53 protein levels, increasing Bcl-2 associated X-protein (Bax) gene expression, and finally enhancing the activities of effector caspases, such as caspase-3.
Based on the present study's results, it was shown that Que, as a natural substance, possesses great abilities in HCC cell apoptosis induction and cancer cell growth reduction. However, the more important results come from the combination of Que and ETO, where according to the present study’s results, it enhances every aspect of ETO’s activities regarding apoptosis induction. Based on the aforementioned results, it is recommended to further investigate the Que + ETO combination as a potentially suitable therapy for HCC in vitro and in vivo.