1. Background
Hepatitis B virus (HBV) is a DNA virus that can be transmitted through body fluids, and epidemiologically, it is considered the main risk factor for hepatocellular carcinoma (HCC). Eight genotypes have been identified for HBV so far (based on > 8% genomic differences in the whole genome) (1). The HBV infection can progress to acute or chronic hepatitis (2, 3). During HBV infection, the viral genome can be integrated into the host’s genome, an important step toward HCC (4). In fact, HBV is considered a major risk factor for HCC via a variety of pathogenic mechanisms, including genomic integration, which can cause genomic instability in the host, alteration in exosomes, autophagy (a modulator of immune responses), and epigenetic changes (5).
Hepatitis B virus infection can adopt a chronic course known as chronic hepatitis B (CHB), which, in its progression, HBeAg is considered a critical marker characterized by long-term HBV replication and modulation of immune responses (6). This condition is a global health problem due to its frequent association with HCC development (7). Although CHB is a notorious condition, it can be controlled by optimal treatment. Actually, the main problem is related to undiagnosed CHB patients who present no clinical manifestation (8). In recent years, the prevalence of HBV has decreased due to vaccination programs, especially in Asia (2). Based on a recent systematic review, 42% of patients with liver cirrhosis globally are infected with HBV (9). Also, occult hepatitis B virus infection (OBI) is another form of CHB and is estimated to affect 0.82% of the general population around the world (10). The prevalence of HBV in Iraq is 2 - 5% in the general population (11), with slight differences in different provinces of the country (11, 12).
Hepatitis B virus displays different genotypes and mutations. Some HBV genotypes are associated with specific mutations, which can affect the biology of the virus and the clinical course of the infection. These variations can enable the virus to escape immune responses, causing treatment resistance and inducing HCC development (13). One of the key determinants of CHB is the core gene of HBV. This gene encodes HBcAg and HBeAg and can undergo multiple mutations. The core protein of HBV is a component of its nucleocapsids, which are necessary for viral DNA reverse transcriptional replication and the attachment of the virus to host cells, ensuring the infection’s protracted course. It is considered that the C gene-encoded protein of HBV is closely associated with its infectivity (14). Mutations in the Pre-Core/Core region can affect the detection of HBeAg and alter the expression of this antigen (15). These mutations can arise due to the lack of proofreading during HBV replication, exaggerating the risk of developing HCC (16) due to the intensified transactivation of this gene by HBx and hepatocyte nuclear factor 1 (HNF1) (17). Another important factor influencing the incidence of Pre-Core mutations and HCC development seems to be the genotype of HBV (18, 19).
2. Objectives
Due to the importance of core protein in the life cycle of HBV, this study aimed to assess HBV Core/Pre-Core mutations and HBV genotypes in patients with chronic HBV in an Iraqi province.
3. Methods
3.1. Patients and Sampling
In the current cross-sectional study, we screened samples from 134 CHB patients for mutations in the Pre-Core/Core gene. All patients who fulfilled our inclusion criteria during the study period were randomly included. Inclusion criteria were serological or molecular confirmed diagnosis of CHB, providing written informed consent, and age >18 years old. Exclusion criteria were failure to obtain enough volume of samples and age less than 18 years old. The diagnosis of CHB was based on having more than 6 months of HBsAg positivity and the presence of the HBV genome in the patient’s serum. Laboratory and demographical data were obtained from patients’ files. The study was conducted after obtaining approval from the Ethics Committee of the Iran University of Medical Sciences (ethical code: IR.IUMS.FMD.REC.1401.221). After obtaining written informed consent, 5 mL of peripheral blood was withdrawn from each patient, and then the serum was separated. The serum sample was stored at -20°C before further analysis for genome extraction. After extraction, DNA samples were stored at -20°C until being used in PCR. We tried to eliminate all sources of bias during the study, including enrollment of patients.
All patients referred to a gastrointestinal and hepatology specialist at Al-Husain Hospital, Kerbala, Iraq, from August to December 2022 were included. The patient’s age, gender, duration of infection, history of receiving treatment, and serum levels of aspartate aminotransferase (AST), alanine aminotransferase (ALT), alkaline Phosphatase (ALP), and albumin (ALB) were recorded by reviewing patient files. Only patients who had no missing data were included in the study.
3.2. Detecting HBV Pre-Core/Core Mutations
All serum samples were subjected to DNA extraction using the AddPrep Genomic DNA Extraction kit (Addbio, South Korea). The extracted DNA was checked using spectrophotometry by reading the 260/280 ratio. The DNA extraction step was repeated for the samples that failed to retrieve adequate amounts of high-quality DNA. Two specific primers were designed to amplify the C gene (Table 1) (more details and the exact location of the primers are provided in Appendix 1, providing the position and length of the retrieved 605 bp and 490 bp amplicons, covering the upstream and downstream portions of the C gene, respectively, within HBV genome (GenBank acc. no. MK693109.1 and LC513656.1). This action could further validate most of the variants identified. Amplification was performed using conventional PCR and AddStart Taq Master (Addbio, South Korea).
| Primers | Sequences (5′ - 3′) | PCR Annealing Temperature (°C) | Amplicon Length (Base Pair) |
|---|---|---|---|
| Primer 1 | 52 | 605 | |
| F | AGGAGGCTGTAGGCATAAATTGG | ||
| R | GTTGTTCTTCTAGGGGACCTGCC | ||
| Primer 2 | 52 | 490 | |
| F | TGCCTTCTGACTTCTTTCC | ||
| R | TGAGGTTCCCGAGAGGGA |
The PCR products were then sequenced following the standard instructions (Macrogen Inc. Geumchen, Seoul, South Korea) by the ABI (Applied Biosystem). Only conclusive chromatographs obtained from sequence files were analyzed. Sequencing results were aligned and analyzed by the reference database and using BioEdit sequence alignment editor software version 7.1 (DNASTAR, Madison, WI, USA). HBV mutations were annotated by SnapGene Viewer ver. 4.0.4 (https://www.snapgene.com). The amino acid sequences of the protein encoded by the target C gene were retrieved from the NCBI server (https://www.ncbi.nlm.nih.gov). The ID of the retrieved core protein with 212 amino acids was QFI36194.1.
3.3. Phylogenetic Analysis
A phylogenetic tree was built to assess the genotypes of the variants identified and their phylogenetic features. All the identified viral variations were grouped according to the pattern of their variations, and specific haplotypes were created for the viral groups identified. DNaSP software, ver. 6.12.01 was utilized to group the haplotype of the variations identified into specific Hap categories (20). A phylogenetic tree was constructed according to the Neighbor-joining method (21). The observed variants were compared with their Neighbor homologous reference sequences available at NCBI-BLASTn (22), followed by visualization as rectangular and circular cladograms by the iTOL suit (23). The sequences of each phylogenetic group in the comprehensive tree were colored appropriately.
3.4. Statistical Methods
The Mann-Whitney U test and chi-squared test were used to analyze the variables, as well as descriptive and analytic statistics, in SPSS version 22 at P < 0.05 as the significance threshold.
4. Results
4.1. Demographical and Clinical Data
The results revealed that 58 (45%) of the patients were male, and 72 (55%) of them were female. The mean age of the patients was 36 ± 12.7 years, and the mean duration of the infection was 5.2 ± 4.8 years. The patients were divided into two groups based on the presence or absence of Pre-Core/Core mutations. There was a significant association between Pre-Core/Core mutations and receiving treatment (chi-square test, P = 0.001) (Table 2).
Abbreviations: AST, aspartate aminotransferase (U/L); ALT, alanine aminotransferase (U/L); ALP, alkaline phosphatase (IU/L); ALB, albumin (g/dL).
a All non-significant P-values are based on the Mann-Whitney U test and the chi-square test.
b Statistically significant based on the chi-square test.
4.2. HBV Pre-Core/Core Mutations
The results demonstrated the presence of 21 nucleic acid variants in the samples investigated in comparison with the reference sequence. These alterations included 111T>C, 138C>T, 204A>G, 300C>A, 306T>C>G, 328T>C, 333C>T, 347T>G, 363A>G, 364A>G, 365T>C, 366A>G, 369C>T, 393T>C, 420G>A, 424T>C, 426A>C, 468G>A, 471C>T, and 544T>C (Appendix 2 and 3). Nucleic acid translation results revealed five amino acid alterations (103G>V, 109I>A, 109I>M, 109I>V, and 110S>A) on the core protein (Appendix 4). These mutations were found in 20% (27 of 134 patients) of our patients.
The NCBI BLASTn engine showed a high similarity (98 - 99% sequence similarity) between 130 sequences evaluated and target sequences with the highest homology (GenBank acc. MK693109.1 related to genotype D). By comparing samples with reference sequences, the accurate position and other details of the PCR fragments retrieved were also identified (Table 3).
| No. | Variant | Position in the PCR Fragment | Position in the Reference Genome | Position in AA | Type of Variant | Variant Summary |
|---|---|---|---|---|---|---|
| (A) Variations Based on GenBank MK693109.1 | ||||||
| 1 | 111T>C | 111 | 1887 | 24L | Silent | 24L |
| 2 | 138C>T | 138 | 1914 | 33D | Silent | 33D |
| 3 | 204A>G | 204 | 1980 | 55S | Silent | 55S |
| 4 | 300A>C | 300 | 2075 | 87A | Silent | 87A |
| 5 | 306T>C>G | 306 | 2082 | 89L | Silent | 89L |
| 6 | 328T>C | 328 | 2104 | 97L | Silent | 97L |
| 7 | 333C>T | 333 | 2109 | 98A | Silent | 98A |
| 8 | 347T>G | 347 | 2123 | 103G | Missense | 103G>V |
| 9 | 363A>G | 363 | 2139 | 108P | Silent | 108P |
| 10 | 364A>G | 364 | 2140 | 109I | Missense | 109I>A |
| 11 | 365T>C | 365 | 2141 | 109I | Missense | 109I>M |
| 12 | 366A>G | 366 | 2142 | 109I | Missense | 109I>V |
| 13 | 369C>T | 369 | 2145 | 110S | Missense | 110S>A |
| 14 | 393T>C | 393 | 2169 | 118V | Silent | 118V |
| 15 | 420G>A | 420 | 2196 | 127R | Silent | 127R |
| 16 | 426A>C | 426 | 2202 | 129L | Silent | 129L |
| 17 | 468G>A | 468 | 2244 | 143T | Silent | 143T |
| 18 | 471C>T | 471 | 2247 | 144V | Silent | 144V |
| 19 | 544T>C | 544 | 2320 | 169L | Silent | 169L |
| (B) Variations Based on GenBank LC513656.1 | ||||||
| 20 | 300C>A | 300 | 2074 | 87A | Silent | 87A |
| 21 | 424T>C | 424 | 2198 | 129L | Silent | 129L |
4.3. HBV Genotypes and Phylogenetic Analysis
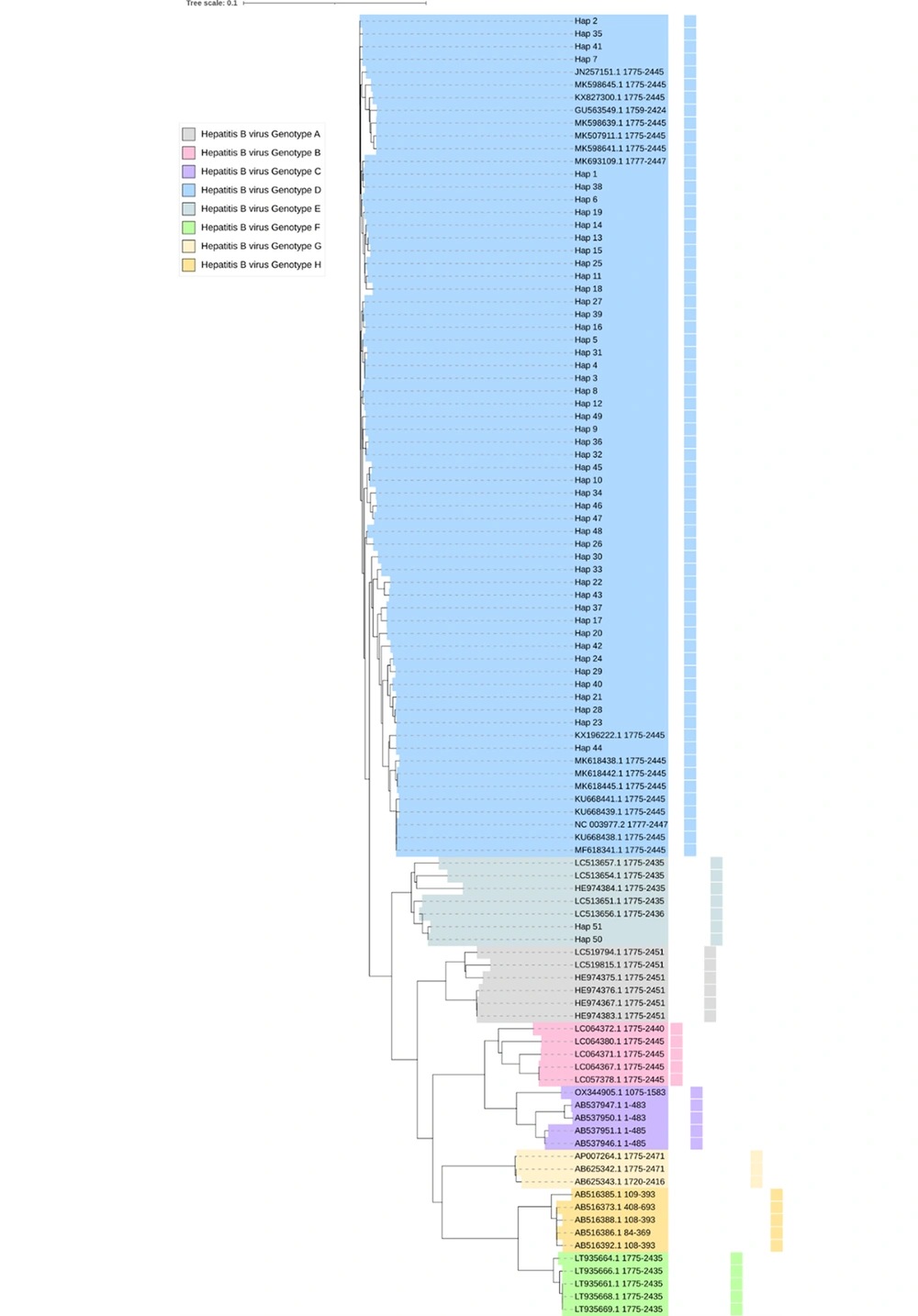
In the phylogenetic tree obtained, samples were divided into 2 genotypes. Most samples were suited in the genotype D clade, and only 4 samples were positioned adjacent to the genotype E clade. The haplotyping of all identified clinical variations of HBV in the studied populations has been investigated. Due to the large sample sizes of the samples investigated, one haplotype was generated for each identified nucleic acid variation to reduce the number of samples incorporated into the phylogenetic tree. Accordingly, 51 haplotypes were generated for all viral samples, 49 of which were attributed to the main group, and two haplotypes were attributed to the minor group (Appendix 5). Aside from the two genotypes reported in this study, six clades were positioned to represent genotypes A, B, C, G, H, and F, respectively (Figure 1 and Appendix 6).
The comprehensive rectangular cladogram of the phylogenetic tree of genetic variants in the C gene of 134 hepatitis B virus genomes extracted from infected humans. All the numbers provided in this image refer to GenBank accession numbers of respective species. The number “0.1” at the top of the tree refers to the degree of the scale range among comprehensive tree-categorized organisms. The letter “Hap_#” refers to the haplotype of the samples investigated.
5. Discussion
As a worldwide health issue, CHB poses a major risk factor for HCC development. The C gene’s mutations in HBV due to the lack of proofreading during DNA replication exaggerate the risk of HCC due to the enhanced activation of HBx and HNF1 (16, 17). In the current study, we evaluated Pre-Core-Core mutations in CHB patients in an Iraqi province. The results revealed 21 nucleic acid variants corresponding to five amino acid substitutions (103G>V, 109I>A, 109I>M, 109I>V, and 110S>A) in the core protein’s amino acid sequence. Hussain et al. (24), in a study in Iraq, identified Pre-Core/Core mutations in 50% of CHB patients, regardless of the HBeAg status, and most of their samples belonged to genotype D. In a study from Iran, Farshadpour et al. (25) detected Pre-Core/Core mutations in 11 out of 19 diabetic patients carrying HBV DNA, most of which were also genotype D (subtypes D1 and D3). In terms of more frequent mutations (such as alterations in amino acid residues 109 and 143 in the Pre-Core/Core protein), our findings are similar to those of Farshadpour et al. In addition to genotype D, Pre-Core/Core mutations are also found in other HBV genotypes as well (26).
None of the mutations identified in our study were associated with HBeAg negativity (27). Amino acid substitutions at positions 109 I>A (nucleotide 2140) (28) and 109I>M (nucleotide 2141) (29) have been reported in a previous study, which seems to be clinically insignificant. Meanwhile, a mutation at the nucleotide position 2142 (109 I) (109I>V in our study) was reported to be associated with HCC development (30). Furthermore, alterations at the nucleotide position 2145 (110S) (110S>A in our study) seem to be more frequent in CHB patients. Our study revealed that 20% of our samples carried Pre-Core/Core mutations. Differences observed in the prevalence of Pre-Core/Core mutations can be due to different sample sizes and geographical distribution of CHB in Iraqi provinces. The exact amino acid substitutions and corresponding mutations observed in our study have also been reported by most previous studies. The low frequency of these amino acid alterations could be due to the small sample size of our study.
The phylogenetic tree drawn clustered our samples into eight clades, indicating that the C gene’s sequences had closer phylogenetic positions in genotypes D (where most of our samples were positioned) and E (where four of our samples were positioned) compared to other genotypes.
In our phylogenetic tree, genotype E was positioned next to genotype D. In addition to the major clades of genotypes D and E, a number of other clades related to other genotypes were also identified. Next to the genotype E clade, six clades representing genotypes A, B, C, G, H, and F were positioned. Our observation confirmed that genotype C was closest to genotypes E and D, respectively. However, the sequences of the C gene-based G genotype have been less frequently deposited in the NCBI database. It appears that genotype D is the main HBV genotype in Iraq (24, 25, 31). There are some reports suggesting a role for HBV genotype in HCC development. However, due to the complex pathogenesis of HCC, the exact role of the HBV genotype in tumorigenesis is not clear, but this issue remains an important topic for future research (18, 19). Furthermore, it was inferred from the phylogenetic tree obtained here that the sequences of genotype D occupied ancestral positions compared with other viral clades related to genotypes E, A, G, B, C, H, and F, respectively.
We need to mention some limitations of our study, including the small sample size and unavailability of the HBeAg status. Further studies with larger sample sizes and more diversified groups of patients are suggested. The HBeAg status in CHB patients carrying Pre-Core/Core mutations can help resolve diagnostic challenges in these patients. In addition, following up on patients for complications such as fibrosis, cirrhosis, and HCC is highly recommended in future studies. Other limitations include short periods of patient recruitment and follow-up. Long-term follow-up of patients can provide a perspective on the clinical significance or outcome related to Pre-Core/Core mutations in Iraqi HBV patients. It is noteworthy that this study provides a primary report on the frequency of Pre-Core/Core mutations in CHB Iraqi patients and highlights the importance of this issue, evidenced by the high prevalence of these mutations in the studied population. Another limitation of this study was the fact that we did not evaluate other HBV genes (such as the S or X genes) or even the host’s genes, whose mutations may be associated with Pre-Core/Core mutations.
5.1. Conclusions
We detected 21 nucleotide variants and five respective amino acid substitutions within the C gene of the HBV genome in Iraqi patients. The mutations detected in the C gene were more frequent in patients who had not received treatment. Genotype D was the main genotype identified, probably highlighting the importance of this genotype in HBV infectivity in our country. Only four samples belonged to genotype E. Most of the variants identified led to noticeable changes in the evolutionary positioning of HBV. However, all these variations did not deviate from the main genotype. The identification of these mutations in the C gene underlies the importance of pursuing these mutations in CHB patients. Future studies are advised to investigate C gene mutations in larger numbers of CHB or HCC patients so that we can draw more conclusive interpretations of the clinical implications of these mutations.