1. Background
Hepatitis B virus (HBV) infection affects more than 250 million people globally (1). This systematic review and meta-analysis aimed to determine the prevalence of HBV infection among hemodialysis (HD) patients worldwide. The results indicated that the pooled prevalence of HBV infection among HD patients was 7.32% (95% CI: 6.53 - 8.15%; I2 = 97.91%) (2). HBV has the potential to progress to liver cirrhosis and cancer, causing nearly one million deaths annually due to liver-related complications, with treatments demonstrating limited efficacy (10 - 30%) (3, 4). The precise cause of HBV infection chronicity remains unclear. Nevertheless, factors such as viral load, age, duration of infection, and host genetics may exert significant influence (5).
In HBV infection, the T cell response has been recognized as playing a crucial role. Weakness in T cell response is a common condition in patients with HBV, influenced by various factors (3). Regulatory T cells (Treg) represent a subset of CD4+ T lymphocytes that regulate immune balance through direct cell contact or the secretion of cytokines. Forkhead box P3 (FoxP3), a transcription factor, serves as a major regulator for Treg function and development (6, 7).
FoxP3 belongs to the Fox protein family and consists of 11 exons encoded in the Xp11.23 region (8). Hepatitis B virus infection significantly increases FoxP3 expression, promoting virus replication and stability by elevating Treg frequency (9).
The FoxP3 −3279 (rs3761548) C/A polymorphism resides in the promoter region of the FoxP3 gene and serves as a binding site for the transcription factor specificity protein 1 (Sp1). Polymorphisms in FoxP3 can modulate its expression levels, impacting Treg-suppressive capabilities (10). An allele of this polymorphism is associated with reduced FoxP3 expression. Extensive research has explored the association of this polymorphism with various human diseases (11).
2. Objectives
This study investigates the impact of the FoxP3 -3279 polymorphism and its expression on chronic HBV infection, recognizing the significant role of host genetics in influencing HBV infection chronicity.
3. Methods
3.1. Samples
This is a case-control study conducted in Tehran in 2023. The study comprised 140 healthy individuals without infections with HCV, HBV, or HIV viruses, serving as controls, and 70 patients with chronic HBV infections as cases. These samples were retrieved from a previous study and stored at -40°C. They were also sourced from the Keyvan Virology Laboratory in Tehran. The study was carried out at the Department of Immunology and Microbiology at the University of Alzahra.
3.2. DNA Extraction
Genomic DNA was extracted from 100 microliters of buffy coat using a commercially available kit (Sinapure DNA-Sinaclon kit) following the manufacturer's instructions. The extracted DNA was stored at -20°C for genotype analysis. The quantity of purified DNA was assessed using a spectrophotometer (nanodrop).
3.3. FoxP3-3279C/A Gene Polymorphism
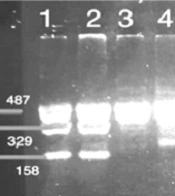
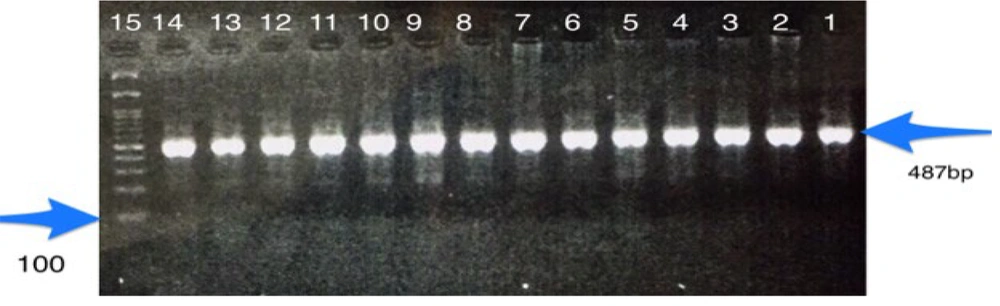
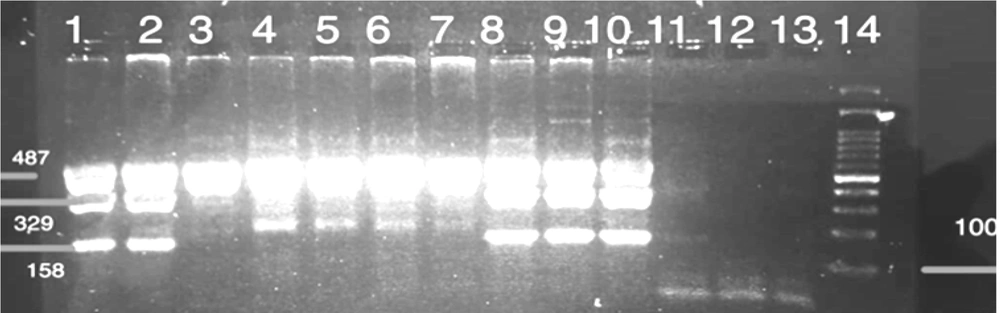
The FoxP3-3279 C/A gene polymorphism was determined through restriction fragment length polymorphism-PCR (PCR-RFLP). For this purpose, the primer sequences listed in Table 1 were employed. The final PCR volume was 20 µL, comprising 1 µL of DNA (100 ng/µL), 10 µL of Master Mix (Amplicon), 1 µL of each primer (10 µM) (SinaClon, Iran), and 6 µL of PCR Grade Water. The thermal program used for PCR amplification included an initial denaturation at 95°C for 5 minutes, followed by 35 cycles of denaturation at 95°C for 30 seconds, annealing at 55°C for 30 seconds, and extension at 72°C for 30 seconds. Finally, a final extension was performed at 72°C for 10 minutes. The PCR products were then electrophoresed on a 2% gel to confirm the results. A 487 bp band for PCR products was digested by Pst1 enzyme in 20-µL volumes as follows: 1 µL of restriction enzyme (1 U/µL), 2 µL of 10 × Buffer, 7 µL of Nuclease-Free Water, and the 10 µL PCR product was incubated at 37°C for 16 hours in a water bath. The fragments were subsequently electrophoresed on a 2% gel to determine the genotype. The genotype included three different patterns of bands: AA (487 bp), AC (487 bp, 329 bp, and 158 bp), and CC (329 bp, 158 bp).
| SNP | rs Number | Primer Sequence | Tm | %GC | Reference |
|---|---|---|---|---|---|
| FoxP3-3279 C/A | 3761548 | F- 5’- GCCCTTGTCTACTCCACGCCTCT-3’ | 66 | 60/87 | (12) |
| R-5’- CAGCCTTCGCCAATACAGAGCC-3’ | 63.98 | 59/09 |
3.4. Real-Time PCR
To assess the differences in FoxP3 gene expression between patients and healthy individuals with different gene polymorphisms, we selected 25 healthy samples as controls and 25 patient samples as cases. Total RNA was extracted from 200 microliters of blood samples (Buffy Coat) using a Roche kit. Subsequently, cDNA synthesis was performed utilizing a Pars Toos cDNA synthesis kit with specific primers (see Table 2), and the GAPDH gene was used as a reference gene for normalization. The final volume for Real-Time PCR (15 µL) consisted of the following components: 7.5 µL SYBR green Master Mix (without ROX), 1 µL of each primer (SinaClon, Iran), 2.5 µL cDNA, and 3 µL DEPC water. The Real-time PCR was conducted using the Rotor-Gene Q System.
| Gene and Primer Sequence | Product Size (bp) |
|---|---|
| FoxP3 | 79 |
| F: 5’CAGCACATTCCCAGAGTTC3’ | |
| R: 5’CGTGGCGTAGGTGAAAG3’ | |
| GAPDH | 107 |
| F: 5’CTTCCAGGAGCGAGATCCCT3’ | |
| R: 5’AATGAGCCCCAGCCTTCTC3’ |
3.5. Statistical Analysis
The data were analyzed using IBM SPSS 26.0 software. Genotype and allelic polymorphism frequencies between the control and case groups were compared using the χ2 test. Additionally, Real-time data were analyzed using REST 2009 software. A P-value < 0.05 was considered statistically significant.
4. Results
4.1. Specification of Study Groups
The control group comprised 130 (92.9%) males and 10 (7.1%) females with an average age of 37.71 ± 9.17. The case group included 68 (97.1%) males and 2 (2.9%) females with an average age of 38 ± 8.41. There were no significant differences in age and sex between the case and control groups (P > 0.05).
4.2. Genotype and Allele Frequencies of the FoxP3-3279 Polymorphism
The study's results displayed a 487 base pair band for the FoxP3 gene polymorphism, as shown in Figure 1. Figure 2 exhibited three distinct band patterns: AA, AC, and CC for the FoxP3-3279 polymorphism. It was determined that this polymorphism was in Hardy-Weinberg equilibrium (P > 0.05) for both the case group (P = 0.200) and the control group (P = 0.140).
The study also analyzed the results based on allelic and four hereditary models (Table 3). The overdominant model suggested that the AC genotype significantly protects against HBV infection compared to CC+AA genotypes of the FoxP3-3279 polymorphism (OR = 0.167; 95% CI = 0.074 - 0.375; P = 0.000), whereas the codominant model indicated that the mutant genotype increases the risk of HBV infection compared to the wild CC genotype (OR = 0.406; 95% CI = 0.194 - 0.849; P = 0.017).
| Variables and Inherited Models and Genotype | Control Group, No. (%) | Case Group, No. (%) | P-Value | OR (95% CI) |
|---|---|---|---|---|
| Codominant | ||||
| CC | 17 (12.1) | 25 (35.7) | - | Reference |
| AC | 61 (43.6) | 8 (11.4) | 0.000 | 0.089 (0.034 - 0.233) |
| AA | 62 (44.3) | 37 (52.9) | 0.017 | 0.406 (0.194 - 0.849) |
| Dominant | ||||
| CC | 17 (12.1) | 25 (35.7) | - | Reference |
| AC+AA | 123 (87.9) | 45 (64.3) | 0.000 | 0.249 (0.123 - 0.503) |
| Overdominant | ||||
| CC+AA | 79 (56.4) | 62 (88.6) | - | Reference |
| AC | 61 (43.6) | 8 (11.4) | 0.000 | 0.167 (0.074 - 0.375) |
| Recessive | ||||
| CC+AC | 78 (55.7) | 33 (47.1) | - | Reference |
| AA | 62 (44.3) | 37 (52.9) | 0.242 | 1.411 (0.793 - 2.509) |
| Allelic | ||||
| C | 95 (92.1) | 58 (37.9) | - | Reference |
| A | 185 (69.3) | 82 (30.7) | 0.133 | 0.726 (0.478 - 1.102) |
4.3. Real-Time PCR
The first step involved checking melting curves for each sample of the target gene (FoxP3) and the reference gene (GAPDH) at temperatures of 85.5 and 87°C, respectively. Amplification and melting curves for both genes were examined to ensure the accuracy of the reaction. Ct values were recorded for each sample, for both the target and control genes. The PCR reaction efficiency for the FoxP3 and GAPDH genes was determined to be 0.9. The results were analyzed using REST software, which revealed that the expression of the FoxP3 gene was significantly increased by 92.3 times (95% CI 92.302 - 92.302), compared to the reference gene (GAPDH gene), which was considered as 1 (P = 0.00).
5. Discussion
Regulatory T cells, which regulate the function of various immune cells, including CD4+ T cells, CD8+ T cells, effector T cells, and antigen-presenting cells, can contribute to persistent HBV infection and disease progression (10, 13). Our study revealed that the FoxP3 -3279 AC (rs3761548) genotype can play a protective role against HBV infection when compared to AA+CC genotypes (P = 0.000).
Conversely, the mutant AA genotype was more prevalent than the AC+CC genotypes in the case group (P = 0.017). The FoxP3 -3279 AA (rs3761548) genotype leads to a loss of binding to E47 and c-Myb transcription factors, resulting in low protein expression (10). The reduction in FoxP3 protein levels is associated with decreased Treg-suppression function (14).
There is limited research regarding the association of FoxP3 (+) regulatory T cells gene polymorphism (rs3761548) with chronic hepatitis B virus infection. In this study, the frequency of the AA genotype in the case group was 52.9%, compared to 44.3% in the control group with CC+AC genotypes, which was not statistically significant (P = 0.242). This finding aligns with a study in Turkey that showed a higher prevalence of the AA genotype in infected individuals, though not statistically significant (15).
It has been observed that the AA genotype of this specific polymorphism increases the risk of colorectal (16) and prostate cancers (17). Additionally, it has been associated with tumor progression in breast cancer. FoxP3 is suggested to have a dual function in tumor suppression and immunomodulation, making it challenging to determine the predominant function in disease progression (18).
A meta-analysis study has indicated that a specific allele of this polymorphism is associated with autoimmune disease (AD) (19). In patients with gastric adenocarcinoma (GA), AA+AC genotypes have been linked to disease progression, and it has been noted that the levels of IL10 and IL35 are significantly higher in patients with the AA genotype (20).
Similarly, a study on human papillomavirus (HPV) infection has shown that the AA genotype is protective against HPV infection (21). Therefore, further research is necessary to determine the impact of mutant AA and AC genotypes on the factors influencing the chronicity of HBV infection, as well as to understand the role of this polymorphism in other viral infections.
Our research has demonstrated that individuals with HBV infections exhibit significantly higher expression of FoxP3 (P = 0.000), which can influence Treg cells and contribute to viral DNA replication, leading to chronicity. Other studies have also reported increased expression of both FoxP3 and Treg cells in HBV infection, with a correlation between Treg frequency and viral load. Regulatory T cells have also been implicated in the progression of hepatitis B infection (9, 22). It is important to investigate the role of FoxP3 -3279 (rs3761548) polymorphism in immunotherapy, as Treg cells play a significant role in immunotherapy (23), and the effect of this polymorphism on Treg cells function needs to be further investigated. There are changes in GAPDH expression in patients with either cirrhosis or cancer, but there is a limitation in this study because it lacks information on the cirrhosis or cancer status of these patients.
5.1. Conclusions
Our research indicates that the FoxP3-3279 AC genotype may offer protective effects against HBV infection when compared to the AA and CC genotypes. Furthermore, we observed a significantly higher prevalence of the AA genotype among individuals with chronic HBV infection in contrast to those with the wild-type CC genotype. However, further investigations are essential to establish the relationship between FoxP3 gene polymorphisms and Treg cell function, as well as to assess the impact of these polymorphisms on the outcome of HBV infection.