1. Introduction
Acute hepatitis of unknown etiology refers to liver injury caused by the exclusion of known infectious or non-infectious factors (1). This condition presents symptoms such as jaundice, vomiting, lethargy, diarrhea, abdominal pain, fever, nausea, and, in some cases, respiratory symptoms (2). Following the notification on April 5, 2022, by the United Kingdom regarding the identification of severe acute hepatitis cases in children under 16 years of age, the World Health Organization (WHO) expressed concern in its technical note dated April 23 about the possibility of an epidemic. The WHO also reported that the cases had no obvious epidemiological risk factors, such as recent international travel, and most cases had not received the COVID-19 vaccine (3, 4). The cause of the hepatitis is currently unknown, but a viral infection link has been suggested due to the epidemiological pattern of the cases (5). Non-A to E hepatitis virus infections were proposed as the possible causal pathogen, as well as a complex mix of genetic risk factors representing challenges for a correct understanding of infectious disease. In these children, no specific pathogen could be proposed as the causal agent (6).
Other theories have been proposed, such as its association with SARS-CoV-2 or an inappropriate response to superantigens, but no mechanism has yet been established (7, 8). Most reported cases did not appear to be epidemiologically linked, and the etiology of this severe acute hepatitis remains unknown and under investigation. The global risk level is currently assessed as moderate (1, 9). We present a case report of a pediatric patient exhibiting the characteristics of acute hepatitis of unknown etiology.
1.2. Sequencing and Bioinformatic Methods
1.2.1. Sample Sequencing
RNA extraction was carried out using the AllPrep DNA/RNA Micro Kit following the manufacturer's instructions, starting with a liver biopsy. Reverse transcription of the hexamer-primed RNA fragments was conducted using the Illumina Stranded Total RNA Prep library kit to produce the first-strand complementary DNA. The RNA template was then removed, and a replacement strand was synthesized to generate blunt-ended, double-stranded cDNA fragments. This process involved PCR amplification of the anchor-ligated DNA fragments, incorporating indexes and primer sequences for cluster generation. The resulting product was a dual-indexed library: DNA fragments with adapters at each end, ready for sequencing using a NextSeq2000 Illumina sequencer.
1.2.2. Bioinformatic Analysis
Sequencing adapters were trimmed, and low-quality sequencing reads were eliminated using Trimmomatic (10). Reads were aligned to the human reference genome hg38 using Burrows-Wheeler Aligner (BWA) (11). Unaligned reads were extracted using samtools (FLAG 0×4) and converted to FASTA format. These reads were then aligned to the KRAKEN bacvir_k31 database, which contains all reference bacterial and viral genomic sequences (12). KRAKEN split each read into subsequences (kmers) of a specific size (size = 31) and searched for each kmer in the bacvir_k31 database. KRAKEN attempted to classify each read into the deepest taxonomical label possible based on its kmers’ classification. Only reads with no unidentified kmers were further analyzed (excluding sequences with the tag '0:' for any of its kmers). Bacteria and viruses associated with more than 500 reads were selected, and their reference genomes were downloaded from NCBI to create a custom BWA index. All non-human reads were aligned against this custom index using BWA.
2. Case Presentation
A previously healthy 4-year-old boy with an incomplete vaccination history presented with symptoms of vomiting, jaundice, and dark urine. Fifteen days later, he experienced nosebleeds and was taken to the emergency room. Physical examination revealed significant jaundice in the sclerae and integuments, hepatomegaly (6 × 3 × 2 cm), without splenomegaly.
A hepatobiliary ultrasound showed signs of hepatic inflammation, while an abdominal CT scan indicated hepatosplenomegaly and intraperitoneal free fluid, with no evidence of parenchymal lesions, obstruction, or bile duct dilation. Serum levels of aspartate aminotransferase and alanine transaminase were both above 500 IU/L. Laboratory tests conducted at various time points during treatment are detailed in Table 1, indicating persistently elevated levels of these enzymes. The patient also exhibited anemia, thrombocytopenia, and reduced hemostatic capacity, suggestive of liver damage as evidenced by increased bilirubin and pancreatic enzyme levels. Initial treatment included omeprazole (1 mg/kg/day), lactulose (0.7 g/kg/day), and vitamin K (10 mg/day).
| Parameter/Date | 06.05.22 | 09.05.22 | 16.05.22 | 24.05.22 | 26.05.22 | 31.05.22 |
|---|---|---|---|---|---|---|
| Hematic biometry | ||||||
| Hemoglobin/Hematocrit, g/dL/% | 10/31 | 10.9/32 | 10.1/33.4 | 11.7/36.9 | 13/42 | 11.6/36.7 |
| White blood cell, u/L | 2750 | 2410 | 2740 | 2050 | 5870 | 4050 |
| Neutrophils, %/u/L | 65.8/1810 | 64/1540 | 77.8/2140 | 41.9/1860 | 84/4940 | 61.3/2480 |
| Lymphocytes, %/u/L | 17.2/470 | 19.6/470 | 13.2/360 | 47.7/960 | 9/530 | 22.5/910 |
| Neutrophils segmented, %/u/L | 4.9/130 | 3.7/90 | 2.3/60 | 2.5/50 | 1.3/70 | 2.3/90 |
| Platelets, u/L | 208000 | 229000 | 277000 | 198000 | 229000 | 444000 |
| Coagulation time | ||||||
| TP/TPT, seg | 28.3/40.6 | 13.1/29.8 | 12.9/31.8 | 12.6/29.2 | 11.3/23.6 | |
| INR | 2.48 | 1.13 | 1.11 | 1.09 | 0.97 | |
| Blood biochemistry | ||||||
| Albumin, g/dL | 3.4 | 3.6 | 3.9 | 4.1 | 4.3 | |
| Creatinine, mg/dL | 0.36 | 0.27 | 0.29 | 0.17 | 0.35 | 0.3 |
| Glucose, mg/dL | 71 | 77 | 123 | 66 | 81 | 73 |
| Ureic nitrogen, mg/dL | 7 | 10 | 9 | 4 | 14 | |
| Calcium, mg/dL | 9.3 | SR | 9 | 9.5 | 10.4 | |
| Chloride, meq/L | 107 | 106 | 103 | 107 | 106 | 106 |
| Potassium, meq/L | 3.8 | 3.7 | 3.6 | 5.1 | 4 | 4.3 |
| Sodium, meq/L | 138 | 137 | 138 | 141 | 140 | 141 |
| Phosphorus, mg/dL | 4.0 | 4.1 | 3.4 | 4.4 | 4.5 | |
| Magnesium, mg/dL | 1.76 | 1.93 | 1.89 | 2.15 | 1.95 | |
| Cholesterol, mg/dL | 215 | 212 | 215 | 232 | 308 | |
| Triglycerides, mg/dL | 471 | 525 | 587 | 609 | 705 | 420 |
| Total Protein, g/dL | 5.6 | 5.3 | 5.7 | 6 | 6.3 | 6.4 |
| Ammonium | 100.9 | 86.4 | 105 | |||
| Liver function test | ||||||
| Amylase, UI/L | 47 | 42 | 58 | |||
| Total bilirubin, mg/dL | 17.93 | 17.53 | 18.6 | 14.89 | 13.62 | 6.73 |
| Direct bilirubin, mg/dL | SR | 12.81 | SR | 10.78 | ||
| Lactate dehydrogenase, UI/L | 910 | 568 | 545 | 638 | 367 | 279 |
| Alkaline phosphatase, UI/L | 344 | 260 | 185 | 133 | 160 | 181 |
| Gamma glutamyl trasferase, UI/L | 116 | 92 | 84 | 93 | 129 | 341 |
| Aspartate aminotransferase, U/L | 3404 | 1814 | 2275 | 1813 | 1404 | 581 |
| Alanine transaminase, U/L | 1865 | 1359 | 1643 | 1525 | 1415 | 873 |
Laboratory Findings of Blood Samples
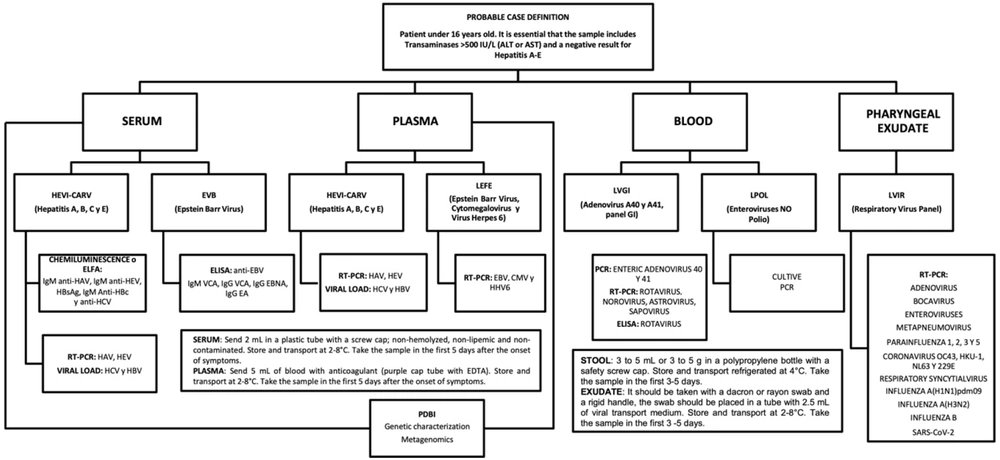
Given that this was the first case of acute hepatitis of unknown etiology in our hospital, a multidisciplinary approach was adopted. This approach was based on the diagnostic algorithm proposed by the Instituto de Diagnóstico y Referencia Epidemiológicos (InDRE), as illustrated in Figure 1 and guided by the “Guide for addressing cases of severe acute hepatitis of unknown cause in children and adolescents" (13).
Workflow based on the sample matrix for investigating acute hepatitis of unknown etiology at the InDRE. For a patient meeting the criteria of a “probable case," the following samples must be collected and corresponding tests conducted to initiate follow-up: Serum: Testing for hepatitis A (HAV), B (HBV), C (HCV), and E (HEV) viruses, Epstein-Barr virus (EBV) IgG, IgM antibodies, anti-SARS-CoV-2, IgG Epstein-Barr nuclear antigens (IgG EBNA), gastrointestinal panel (GIP), and Epstein-Barr premature antigens (IgG EA). Plasma: Testing for HAV, HBV, HCV, and HEV, EBV, cytomegalovirus (CMV), and human herpesvirus 6 (HHV-6). Stool: testing for adenovirus A40 and A41 (LVGI) and non-polio enterovirus (LPOL). Pharyngeal exudate: Respiratory virus panel (LVIR), including bocavirus, enterovirus, metapneumovirus, influenza 1, 2, 3, and 5, coronavirus OC43, HKU-1, NL63, and 229E, syncytial virus, respiratory, influenza A(H1N1) pdm09, influenza A(H3N2), influenza B, and SARS-CoV-2.
According to the diagnostic algorithm, the first step was to rule out infectious etiologies. Therefore, RT-PCR testing for SARS-CoV-2 yielded negative results, along with the absence of anti-SARS-CoV-2 IgM and IgG antibodies in the serum. Furthermore, various samples, including blood, plasma, and stool, were examined for a range of viruses and bacteria, all of which tested negative. These included Adenovirus, Coronaviruses (229E, HKU1, NL63, OC43), Metapneumovirus, Rhinovirus, Enterovirus, Influenza viruses (A, B), Parainfluenza viruses (1-4), Respiratory Syncytial Virus, Bordetella pertussis, Chlamydia pneumoniae, Mycoplasmapneumoniae, Hepatitis A, B, and C viruses, Epstein-Barr Virus, Rotavirus, Astrovirus, enteric Adenovirus, and Mycobacterium tuberculosis.
Immunologists and pediatric hematologists evaluated the patient to rule out primary immunodeficiencies and hemophagocytic syndrome due to persistent bicytopenia. Tests for various antibodies and immune markers, including antinuclear antibodies (ANA), anti-smooth muscle antibodies (anti-SMA), anti-LC-1, anti-SLA, p-ANCAs, anti-mitochondrial antibodies (AMA), anti-LKM-1, IgG, IgA, IgM, C3, C4, α-1 antitrypsin, ceruloplasmin, CD3, CD19, CD8, CD3+CD4, CD3+CD8, CD16+56, CD4/CD8, and direct Coombs, all yielded normal results. Additionally, tumor marker tests for Alpha-fetoprotein (AFP), human chorionic gonadotropin hormone fraction β (β-GCH), and carcinoembryonic antigen (CEA) to rule out malignancy were negative.
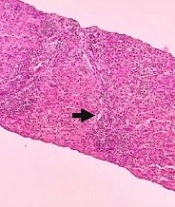
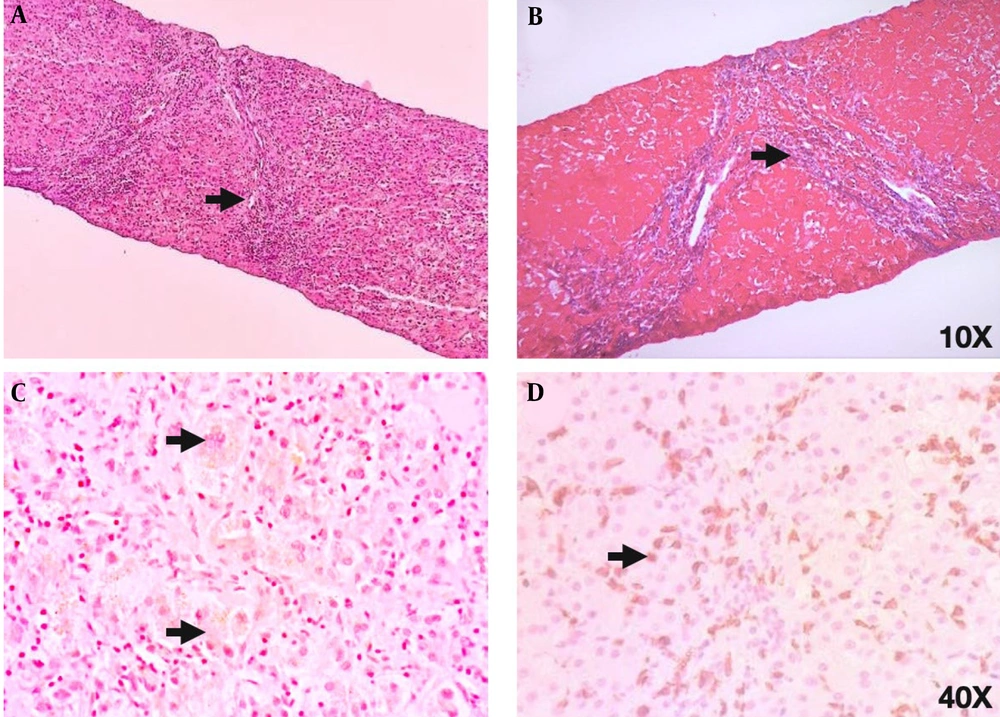
After obtaining negative results from all laboratory tests and observing persistently elevated liver function tests, it was decided to perform a fine needle biopsy of the liver to provide diagnostic support. Histological analysis revealed fibrous tissue with loss of liver tissue, along with a predominance of TCD8+ lymphocytic inflammatory infiltrate and few polymorphonuclear cells exceeding the limiting plaque, suggestive of an active infectious process. Additionally, connective tissue, ductal proliferation, and regenerative changes with multinucleated giant cells were observed (Figure 2).
(A) Hematoxylin and eosin staining: There are 3% of portal spaces with chronic and acute inflammatory infiltrate and interphase activity. The histology reveals the presence of fibrous tissue with loss of liver tissue, thus empty spaces are observed (arrowhead). Additionally, the inflammatory infiltrate suggests an active infectious process. (B) Masson’s trichrome stain shows the presence of fibroconnective tissue (in blue) with duct proliferation in the middle. Liver fibrosis is confirmed by the observation of collagen fibers (arrowhead). (C) Perls stain (PERLS) depicts intracytoplasmic bile in brown color (intracytoplasmic cholestasis) and rules out hemosiderin. A multinucleated giant cell (arrowhead) is observed as a regenerative reactive change. (D) Finally, CD8 positive lymphocytes (brown) (arrowhead) constitute 90% of the observed infiltrate, evident as recruitment during an infectious process.
To identify a potential causative agent of the inflammatory process in situ, high-depth sequencing of the complete transcriptome was conducted. The analysis revealed that out of 15 590 325 sequencing reads, none corresponded to RNA of human origin. These reads were aligned against a database of bacterial and viral genomes. Some reads could be unambiguously attributed to certain bacteria at the species level, while others lacked sufficient information for specific identification. Bacteria and viruses associated with more than 500 reads were selected, and all non-human reads were aligned against the reference genomes of these selected bacteria and viruses.
A total of 3 217 144 reads aligned against these genomes with the following relative abundances: Pseudomonas yamanorum (0.240777742), Stenotrophomonas maltophilia (0.220786344), Clostridium botulinum (0.156878381), Clostridium kluyveri (0.146720602), Novosphingobium ginsenosidimutans (0.099827392), Serratia grimesii (0.079520473), and Citubacterium acnes (0.049570655), along with Human endogenous retrovirus K113 (0.00591841).
To investigate the possibility of a superantigen presence and its potential association with HLA due to a previous asymptomatic SARS-CoV-2 infection, HLA alleles were typed using the SSP and SSO methods, revealing the following alleles: A*11:01, A*24:14; B*15:67, B*38:25; C*O3:53, C*04:169; DRB1* 01:01, DRB1* 16:01, DQB1*03:02, DQB1* 05:01.
As part of the multidisciplinary approach, the gastroenterology service recommended correcting the coagulopathy by continuing the current treatment and adding Rifaximin (30 mg/kg/day orally every 8 hours). Toxicological and ophthalmic factors were ruled out.
After 8 days of hospitalization, the patient exhibited signs of liver failure and was monitored and treated in the pediatric intensive care unit for 9 days without meeting transplant criteria and showing no signs of encephalopathy. Treatment included a hepatopathic diet, anti-ammonium measures, ursodeoxycholic acid (20 mg/kg/day for 12 days), N-acetylcysteine (700 mg for 8 days), and L-ornithine and L-aspartate (for 7 days).
Eight weeks after symptom onset, with minimal clinical improvement and persistently elevated liver function tests, the initiation of immunomodulatory therapy was considered. Treatment began with prednisone at a dosage of 1mg/kg/day. Before commencing treatment, several studies were repeated, including a panel of 13 inflammatory cytokines, which revealed elevated values for IL-1b, IFN-α-2, IFN-γ, TNF-α, MCP-1, IL-6, IL-8, IL-10, IL-12p70, IL-17a, IL-18, IL-23, and IL-33.
Liver function tests decreased to nearly half of the previous levels, along with reductions in IFN-γ, TNF-α, MCP-1, IL-6, IL-8, IL-18, and IL-23. Currently, the patient is undergoing outpatient follow-up with clinical improvement, a reduced prednisone dosage of 0.5mg/kg/day, and vitamin supplements (A, C, and D), with close monitoring of transaminases, coagulation times, hemoglobin, and leukocyte count.
3. Discussion
The issue of acute severe hepatitis of unknown etiology in children has received considerable attention within the scientific community, particularly since early 2022. A variety of potential causes and contributing factors have been investigated, reflecting the complexity of this condition.
Acute severe hepatitis of unknown etiology in children has emerged as a significant diagnostic and therapeutic challenge. Our report underscores the complexity of this condition, suggesting immunological mechanisms and dysbiosis of intestinal microbiota as contributing factors. These findings align with recent research indicating a connection between gut microbiota and liver diseases (14, 15).
Initially, the role of various pathogens in the development of this condition has been explored. Detection of enterovirus, parechovirus, human herpesvirus 6 and 7 (HHV-6, HHV-7), varicella-zoster virus, and adenoviruses in certain cases suggests a potential viral involvement. In Alabama, for example, six out of nine cases tested positive for Epstein-Barr Virus (EBV) by PCR. Other identified viruses include enterovirus/rhinovirus, metapneumovirus, respiratory syncytial virus, and human coronavirus OC43. Preliminary metagenomics findings have also indicated the presence of Adeno-associated virus 2 (AAV2), Adeno-associated dependoparvovirus A, Human Herpes Virus, and Human Polyomavirus in samples from England and Scotland (16).
Moreover, the detection of HERV-K113 in our patients introduces new avenues for research. While its precise role in acute hepatitis remains unknown, previous studies have linked endogenous human retroviruses with autoimmune diseases (17, 18). This supports the hypothesis that HERV-K113 could contribute to pathogenesis by promoting inflammation and altering the immune response.
Regarding potential mechanisms, there is a suggestion that intestinal dysbiosis plays a role in influencing hepatic homeostasis and the immune response, which is consistent with current research on the relationship between gut microbiota and liver diseases. The prevalence of certain opportunistic pathogenic bacteria in the gut microbiota has been considered as a potential link to the observed hepatic inflammation in these cases. This hypothesis finds support in previous research indicating that intestinal dysbiosis can trigger inflammation and liver damage (19).
The role of the gut microbiota in the pathogenesis of acute hepatitis is an emerging area of study. Prior research has demonstrated that intestinal dysbiosis can impact hepatic homeostasis and the immune response, leading to inflammation and liver damage (15). In our case, the dominance of specific opportunistic pathogenic bacteria in the gut microbiota hints at a potential association between dysbiosis and the observed hepatic inflammation.
On the other hand, the low genome coverage of the bacteria identified in our sequencing results suggests that their presence might stem from environmental contamination, potentially related to the patient's skin or the instrumentation used during the biopsy procedure. However, even in such cases, our results do not rule out the involvement of a pathological agent in the disease. The pathological agent may not be present in the diseased tissue or may not be included in the reference database utilized in this study.
Regarding the implications of HLA molecular typing, our study did not reveal a clear association with specific alleles. However, previous studies have suggested a link between certain HLA alleles and autoimmune hepatitis (20, 21). It is important to continue investigating the role of genetics in susceptibility to hepatitis of unknown origin.
Our findings emphasize the necessity of considering a wide range of pathogenic factors, including infectious, genetic, and environmental agents, in cases of acute hepatitis of unknown etiology. A comprehensive understanding of these factors is essential for developing effective therapeutic strategies and preventing future outbreaks.
In conclusion, this case underscores the importance of a multidisciplinary approach and the need for further research to fully comprehend the pathological mechanisms of acute hepatitis in children. Future studies should concentrate on exploring the relationship between the gut microbiota, the immune response, and genetics in the etiology of this disease.
3.1. Conclusions
This pediatric case of acute hepatitis with an unknown etiology presented a complex diagnostic challenge. The comprehensive diagnostic algorithm, including extensive laboratory testing, imaging, and liver biopsy, revealed persistent liver inflammation and identified potential contributors such as dysbiosis of the gut microbiota and the presence of Human Endogenous Retrovirus K113 (HERV-K113). Despite ruling out infectious, immunodeficiency, and oncological causes, the exact pathogenic mechanism remains elusive. Furthermore, the identification of HERV-K113 raises questions about its potential role in the pathogenesis, necessitating further exploration.
The clinical course, marked by an initial lack of response to conventional therapies and subsequent improvement with immunomodulatory treatment, underscores the complexity of this case. Elevated inflammatory cytokines prior to treatment initiation and their subsequent reduction suggest a potential link between immune dysregulation and the observed hepatic pathology.
While this study sheds light on possible immune-mediated mechanisms, including dysbiosis and the role of HERV-K113, definitive conclusions require additional research and surveillance. The intricate nature of this case underscores the need for continued vigilance in identifying early signs of similar clinical conditions and warrants further studies to elucidate the precise etiology and optimal management strategies for pediatric acute hepatitis of unknown origin.