1. Background
Liver fibrosis arises from injuries induced by chronic liver damage. During the fibrotic phase, parenchymal cells undergo apoptosis or necrosis, leading to the replacement of damaged tissue with extracellular matrix (ECM) proteins, including collagen (1). Unregulated fibrosis can lead to excessive tissue remodeling, resulting in the formation of permanent scar tissue (2). Untreated liver fibrosis is a major cause of high-mortality liver cirrhosis and hepatocellular carcinoma (HCC) (3). Liver fibrosis is mainly driven by hepatic stellate cells (HSCs) located between hepatocytes and sinusoidal cells (4). In a healthy liver, HSCs secrete ECM components in moderate amounts. However, major tissue damage induces them to convert into proliferative myofibroblast-like cells with no stellate shape, exuding excessive amounts of autocrine activators, which may lead to liver fibrosis (5).
The well-known transforming growth factor-beta (TGF-β) family, comprising more than 30 proteins, is a driving factor for liver fibrosis (6). Transforming growth factor-beta acts through its serine/threonine kinase receptors (TβRI and TβRII), both of which reside across the cell membrane. Following the linkage between TGF-β and its TβRII receptors, TβRI becomes phosphorylated and activates Smad3 (7). Smad3 transduces the signal into the nucleus, affecting various genes and different cell functions, including HSC activation, urging them to secrete more ECM proteins. Thus, increased TGF-β levels correlate with increased liver fibrosis (8).
The post-translational regulatory activities of microRNAs (miRNAs) play important roles in numerous cellular activities, such as cell apoptosis and proliferation (9). microRNAs are short non-coding oligonucleotides, typically consisting of 19-22 bases, that can bind to the untranslated 3' region (3' UTR) of newly transcribed mRNA molecules, thereby inhibiting their translation. This mechanism is significant in the context of liver fibrosis. microRNAs play a dual role, with some being pro-fibrotic and others being anti-fibrotic (10). miR-29b is an anti-fibrotic factor that inhibits HSC activation by down-regulating the TGF-β gene (11). Another well-known anti-fibrotic miRNA is miR-146a, which is down-regulated in liver fibrosis models (12).
In recent times, there has been considerable interest in the potential therapeutic application of mesenchymal stem cells (MSCs) for liver diseases. Mesenchymal stem cells are a heterogenic subtype of pluripotent progenitor cells considered a proper source for tissue rehabilitation (13). The MSCs' multigenerational differentiation capability has drawn attention as a reliable source for treating liver fibrosis (14). However, high production and conservation costs and a relatively long delay before preparation for use are among the challenges of this method.
It has been shown that MSCs secrete extracellular vesicles known as microvesicles and exosomes, both of which are associated with MSC functions by transferring paracrine factors to target cells, promoting tissue regeneration, and regulating immunological responses (15). Exosomes are small vesicles, typically ranging from 30 to 100 nanometers in diameter, that contain various proteins and are secreted by various cell types, including MSCs (16). The utilization of exosomes for therapeutic interventions offers several advantages over conventional cell therapy, warranting a potential shift towards this novel approach. Exosomes derived from MSC cultures not only exhibit greater efficacy but also circumvent the risks and challenges associated with cell therapy, such as transplant rejection (17). Unlike traditional cell therapy, cell injection leads to side effects that are not present in the implementation of exosomes. Moreover, the simplicity and small size of exosomes mean simpler production and preservation (18).
Lipopolysaccharide (LPS), mainly generated by Gram-negative bacteria, is known for inducing pro-inflammatory cells, such as macrophages, and activating pro-inflammatory cytokines, including TNF-α and IL-6. Various reports indicate that the stimulation of immune cells by LPS leads to further secretion of pro-inflammatory factors during inflammation, resulting in more exosomes being generated (19). It is also reported that preconditioned MSCs generate more paracrine effects through producing more exosomes (20). Consequently, ongoing research is investigating exosomes isolated from MSCs, both pre-treated with LPS and untreated, to elucidate their potential impact on liver fibrosis through MSC activation. Currently, our understanding of the functional mechanisms of MSC-derived exosomes in the liver and their effects on liver fibrosis remains limited.
2. Objectives
The present study focuses on the impacts of exosome administration on the anti-fibrotic miR-146a and miR-29b in the liver-based LX2 cell line to determine whether they demonstrate any positive effects on this condition.
3. Methods
3.1. Cultivation and Treatment of HSCs
The immortal HSC cell line, LX-2, was obtained courtesy of Professor S. Friedman. These cells were grown in 6-well plates containing Dulbecco's modified eagle medium (DMEM) supplemented with 10% FBS. Cells were starved and then treated with TGF-β (5 ng/mL) for 24 hours (21). Next, the Wharton's jelly-derived mesenchymal stem cells (WJ-MSCs) exosomes (50 μg/mL) (22), diluted into DMEM, replaced the previous medium and incubated for 24 hours. The experiment consisted of three groups: An untreated control, a TGF-β treated group, and a TGF-β plus WJ-MSCs exosomes treated group. For later experiments, cells were washed twice with phosphate-buffered saline (PBS) and then harvested. The study was conducted with the approval of the Ethics Committee of Ahwaz Jundishapur University of Medical Sciences (IR.AJUMS.REC.1400.461).
3.2. Isolation and Culture of WJ-MSCs
After obtaining consent, fresh umbilical cords from mothers who underwent full-term cesarean deliveries were collected. Following rinsing with PBS, the Wharton's jelly was dissected into small pieces and cultured in DMEM. The umbilical cord segments were then incubated at 37°C.
3.3. WJ-MSCs Differentiation Assays
The research involved conducting experiments on WJ-MSCs in passages 3 - 5. These cells were cultured in media specifically designed for osteogenic and adipogenic differentiation (BN_0012.4 and BN_0012.5, IRAN), respectively. For osteogenic differentiation, WJ-MSCs were seeded at a density of 20,000 cells/mL in a six-well plate and incubated in osteogenic differentiation media for 21 days, with media replacement twice weekly. After the incubation period, the cells were fixed with 10% formalin for 10 minutes and stained with Alizarin Red (Sigma-Aldrich) for 45 minutes at room temperature. For adipogenic differentiation, WJ-MSCs were incubated in adipogenic differentiation media for 21 days, followed by cell staining using 0.5% Oil Red O (Sigma-Aldrich) in methanol for 45 minutes at ambient temperature. Finally, the cells were examined using a confocal microscope.
3.4. WJ-MSCs Surface Marker Identification
After rinsing with PBS, the cells were stained with conjugated monoclonal antibodies according to the manufacturer's instructions. The antibodies used included FITC-conjugated mouse anti-human CD45, PE-conjugated mouse anti-human CD44, PE-conjugated mouse anti-human CD105, and FITC-conjugated mouse anti-human CD34 (all obtained from eBioscience). Cell analysis was performed using the BD FACSLyric instrument (Becton Dickinson, San Diego, CA, USA), recording a minimum of 20,000 events per sample. The results were then analyzed using FlowJo™ software.
3.5. Induction of WJ-MSCs with LPS
To evaluate the effect of LPS preconditioning on MSC differentiation, LPS was diluted in DMEM, and MSCs were treated with a concentration of 1 μg/mL (23). Subsequently, for the differentiation assays, the LPS-containing medium was replaced with serum-free medium.
3.6. Exosomes Extraction
To isolate exosomes, cells were cultured in DMEM supplemented with 15% FBS. Prior to extraction, the FBS concentration was reduced to 0%. The collected medium underwent centrifugation at 300 × g for 10 minutes. Exosomes were then extracted using an EXOCIB extraction kit (CIB Biotech Co., Iran) following the manufacturer's instructions. The isolated exosomes were suspended in 50 - 200 µL of PBS and stored at -80°C. The concentration of exosome-related proteins was determined using a bicinchoninic acid (BCA) kit (Parstous, Iran), with each unit corresponding to the exosome concentration.
3.7. Characterization of Exosome
Exosome size distribution analysis was conducted through dynamic light scattering (DLS). The exosome solution was dissolved in PBS with 0.05% tween-20 to achieve a concentration of 1 μg/mL. The size of the exosomes was determined using the DLS Zetasizer Nano (Malvern Corp, UK) at 23°C, following the manufacturer's guidelines. Visualization of exosomes was carried out using transmission electron microscopy (TEM). To fix the exosomes, a 1% glutaraldehyde solution was applied for 20 minutes. Subsequently, the exosomes were washed with PBS, diluted in distilled water, stained with 1% uranyl acetate, and examined under an electron microscope.
3.8. Quantitative Real-time PCR
Following the isolation of total RNA using the RNA Isolation Kit (Yekta Tajhiz Azma, Iran), cDNA templates were synthesized with a cDNA synthesis kit (Yekta Tajhiz Azma, Iran). Real-time PCR was then conducted using the SYBR Green master mix (Ampliqon, Denmark) and the QuantStudio™ 3 Real-Time PCR System (ABI Applied Biosystems), following the manufacturer's guidelines. The primer sequences were designed by Sinaclon Company (Tehran, Iran) (Table 1). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as a housekeeping gene to normalize expression levels. The 2-ΔΔCT method was used to calculate fold changes.
| Gene | Primers Sequence | Size of PCR Product (bp) |
|---|---|---|
| COLA1 | F. 5′-TGAAGGGACACAGAGGTTCA-3′ | 188 |
| COLA1 | R. 5′-AGGATCATAACCACGACGA-3′ | |
| α-SMA | F. 5′-CAAGTCCTCCAGCGTTCTGA-3′ | 196 |
| α-SMA | R. 5′-GCTTCACAGGATTCCCGTCTT-3′ | |
| Hsa‐miR-146a | F. 5′-UGAGCACUGUAUUCAUCGAU-3′ | 148 |
| Hsa‐miR-129b | F. 5′-UGACAAGUUAAUUCAUGUGU-3′ | 153 |
| U6 | F. 5′-GCAACGCATTTACTAAGGGG-3′ | |
| U6 | R. 5′-ACCGATGGAAGCATTCACGA-3 | |
| GAPDH | F. 5′- TCCCTAGTCAGGAAATGGT-3′ | 181 |
| GAPDH | R. 5′- TTCCAGTTCTCGGATTTGAC-3′ |
Primer Sequence for Reverse Transcription-Polymerase Chain Reaction
3.9. Western Blotting
The content of the phosphorylated Smad3 (p-Smad3) protein was evaluated by western blot analysis. Hepatic stellate cells were lysed using RIPA buffer containing protease and phosphatase inhibitors. Protein concentration was determined using a BCA assay kit (Thermo Fisher Scientific, USA). Protein samples (30 μg per lane) were separated by SDS-PAGE and then transferred to PVDF membranes (Millipore, USA). The membranes were incubated with the primary antibody (1:1000; Cell Signaling, USA) at 4°C overnight, washed three times with TBST, and then incubated with the secondary antibody (1:10,000; Cell Signaling, USA). Detection was performed using an electrochemiluminescence (ECL) detection kit (GE Healthcare, Chicago, IL, USA), and protein bands were visualized using a ChemiDoc device (BioRad). Glyceraldehyde 3-phosphate dehydrogenase was used as a control protein to normalize protein expression levels. Band density was determined using NIH Image J software.
3.10. Statistical Analysis
Each experiment was conducted in triplicate. Data were analyzed using mean ± SEM, and statistical comparisons were made using ANOVA followed by Tukey's HSD tests with GraphPad Prism 9.0 software. A significance level of P < 0.05 was considered statistically significant.
4. Results
4.1. Characterization of WJ-MSCs
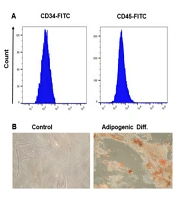
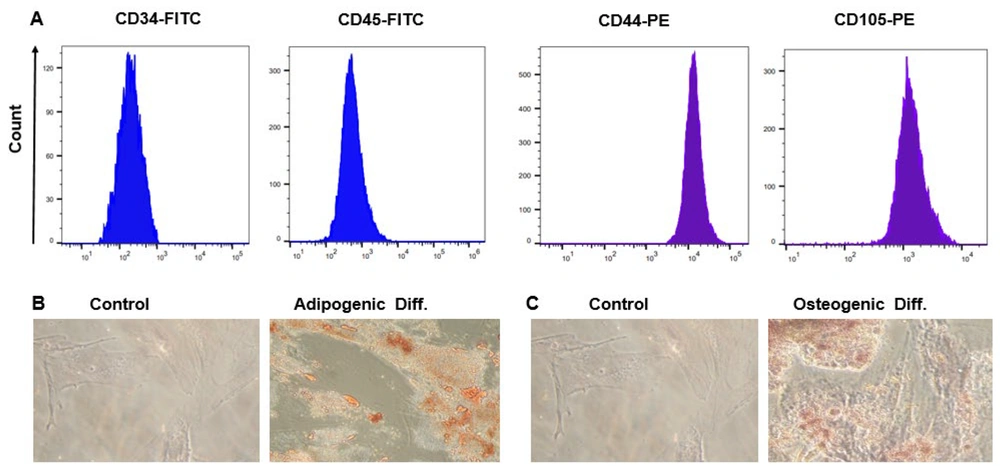
For WJ-MSC characterization, in accordance with the International Stem Cell Association's guidelines, we assessed the surface marker expression and differentiation potential (24). Wharton's jelly-derived mesenchymal stem cells in the third passage formed spindle-like monolayers, resembling fibroblast-like cells adhering to the bottom of the cell culture flasks. Using flow cytometry for surface marker analysis, we found that the MSCs expressed CD44 and CD105 markers while lacking hematopoietic stem cell (CD34) and monocyte-macrophage (CD45) markers. This confirms that only MSCs were isolated and not endothelial or hematopoietic-derived cells (Figure 1A). Oil Red O visualization, after 21 days of culture, demonstrated the formation of numerous lipid droplets, proving the differentiation potential of WJ-MSCs (Figure 1B). Furthermore, inducing cell differentiation into osteoblasts followed by Alizarin Red staining resulted in intense calcium deposition, representing typical osteogenic differentiation (Figure 1C).
Immunophenotyping and differentiation capabilities of Wharton's jelly-derived mesenchymal stem cells (WJ-MSCs) were assessed. A, flow cytometry analysis revealed the expression of CD44 and CD105, while CD34 and CD45 were down-regulated on the surface of WJ-MSCs; B, Oil Red O staining exhibited vibrant red intracellular lipid accumulation in WJ-MSC adipocytes on day 21; C, Alizarin Red S staining showcased intense orange-red staining indicative of calcium deposition in WJ-MSC osteocytes at day 21.
4.2. Exosome Characterization
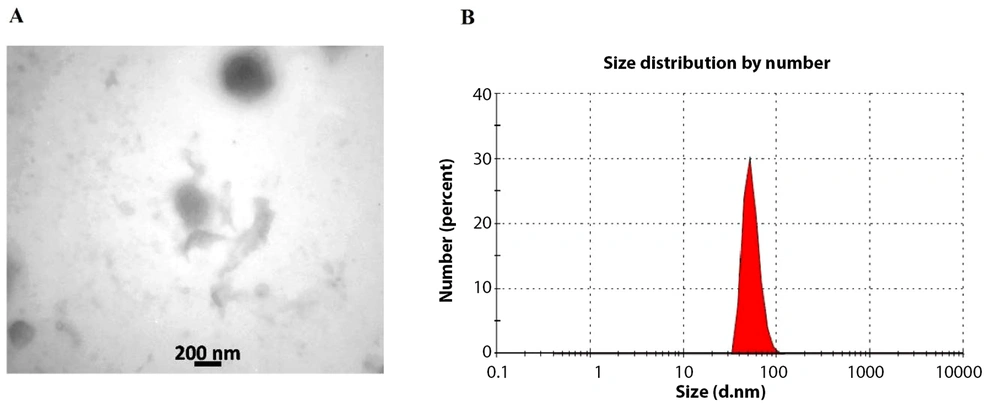
Through TEM, we demonstrated the spherical shape of our exosomes, ranging from 50 - 200 nm in diameter (Figure 2A). The size distribution of the isolated exosomes was determined using a zeta sizer. Approximately 80% of the exosomes had a diameter of 73 nm (Figure 2B).
4.3. Impacts of TGF-β and Exosomes Treatments on Liver Fibrosis Marker Genes
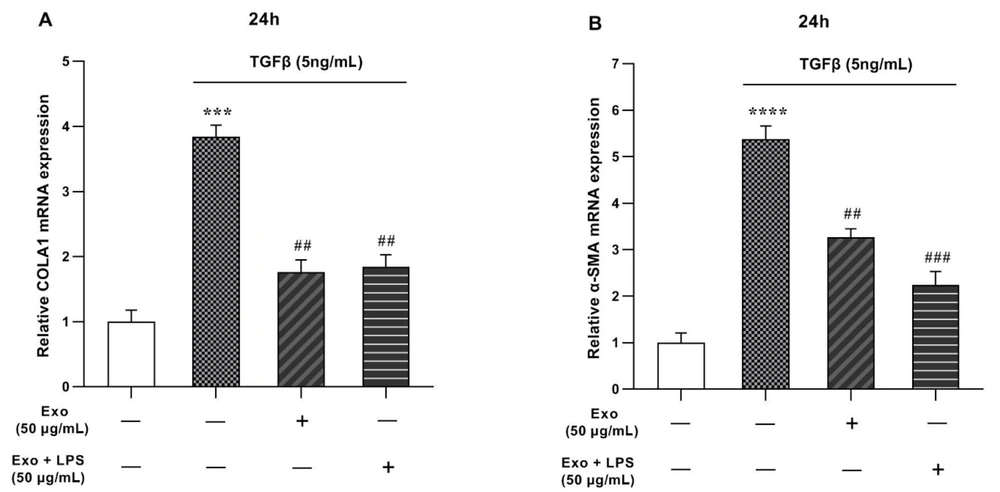
Real-time PCR analysis indicated that TGF-β (5 ng/mL) upregulates markers of liver fibrosis (α-SMA and collagen1α) compared to the control (con) group (Figure 3). The notable increase in mRNA levels of α-SMA and Collagen1α was attributed to liver fibrosis induced by TGF-β. However, when treated with WJ-MSCs exosomes (50 μg/ml), the gene expression of collagen1α and α-SMA declined to 1.76 and 3.26 folds, respectively, even after upregulation by TGF-β (Figure 3A and B). Although the LPS + Exo-treated group showed a significant reduction in α-SMA gene expression compared to the control group, no significant changes were observed in the expression of collagen1α in this group.
mRNA expression levels of collagen type I (COLA1) and alpha-smooth muscle actin (α-SMA) genes in LX2 cell line were evaluated in the presence of transforming growth factor-beta (TGF-β) and exosomes. A, COLA1 gene expression in LX2 cells treated with TGF-β, exosome and, exosome + lipopolysaccharide (LPS); B, αSMA gene expression in LX2 cells treated with TGF-β, exosome and, exosome + LPS. The results, representing three replications (mean ± SEM), were compared to the control. The significance level was set at P < 0.05. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as the reference gene, and statistical significance was denoted as *** P < 0.001, **** P < 0.0001, ## P < 0.01, ### P < 0.001.
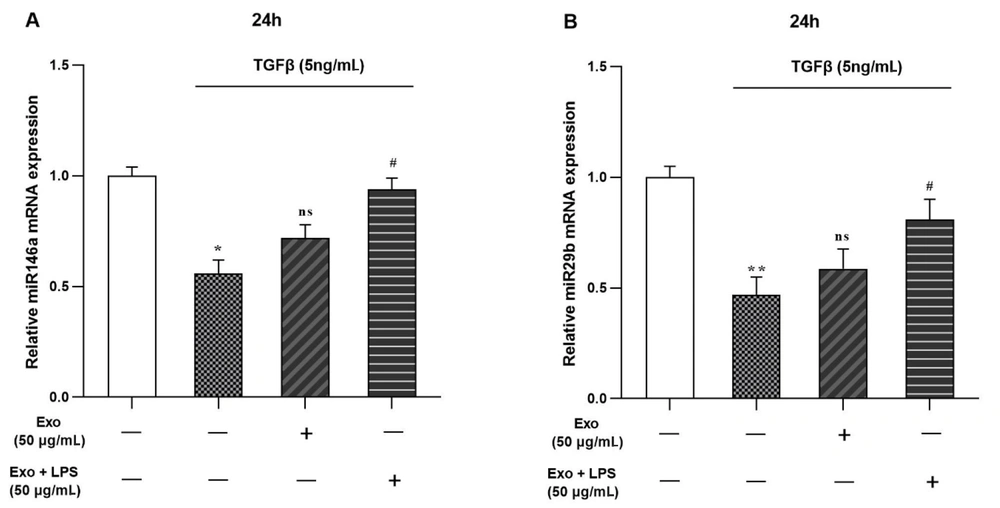
4.4. Effects of TGF-β and Exosomes Treatments on miRs Expression
Our results from the real-time PCR showed that TGF-β (5 ng/mL) treatment leads to the downregulation of miR-29b and miR-146a genes in LX2 cells (Figure 4). Additionally, the addition of WJ-MSCs exosomes (50 μg/mL) did not significantly change their expression. However, after treatment with Exo + LPS (50 μg/mL), both miRs were significantly upregulated to 0.81 and 0.94 folds, respectively (Figure 4A and B).
mRNA expression levels of miR-146a and miR-29b genes were examined in the presence of transforming growth factor-beta (TGF-β) and exosomes in the LX2 cell line. A, miR-146a gene expression in LX2 cells treated with TGF-β, exosome and, exosome + lipopolysaccharide (LPS); B, miR-29b gene expression in LX2 cells treated with TGF-β, exosome and, exosome + LPS. The results, representing three replications (mean ± SEM), were compared to the control. The significance level was set at P < 0.05, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was employed as the reference gene. Statistical significance was denoted as * P < 0.05, ** P < 0.01, # P < 0.05.
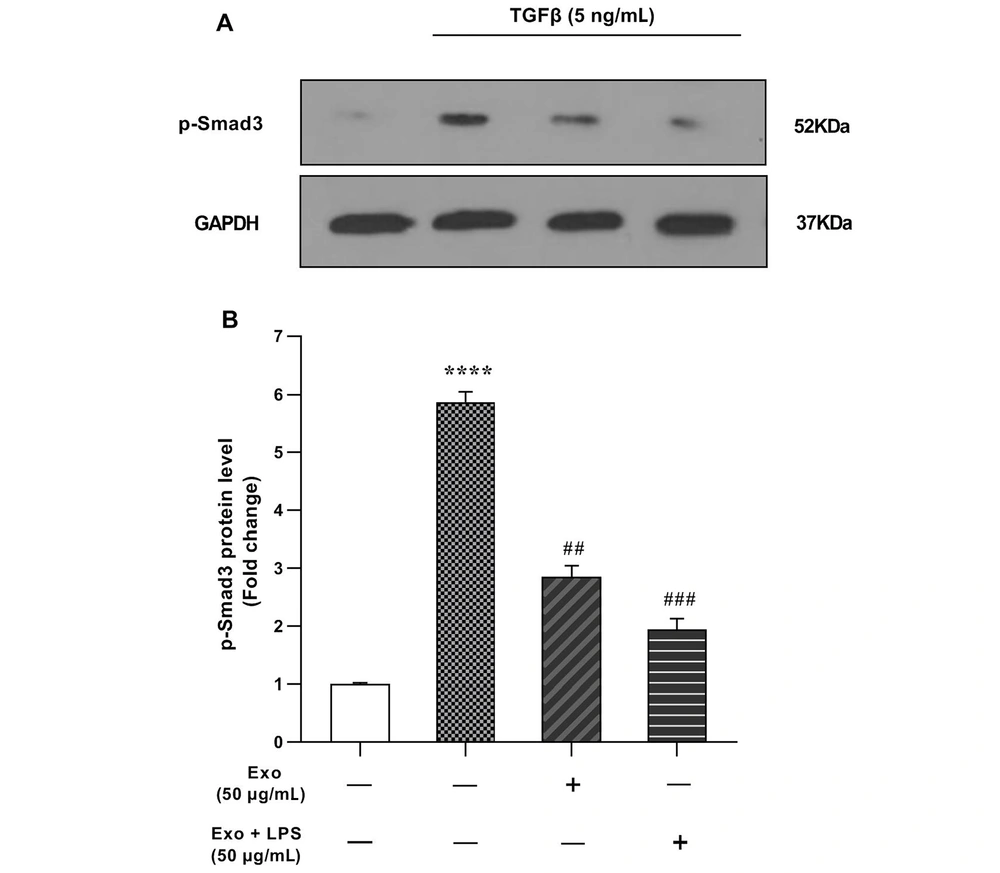
4.5. Effects of TGF-β and Exosomes Administration on Smad3 Phosphorylation
We investigated the impact of exosomes on the TGF-β/Smad3 signaling in LX2-HSC cells. After 1 hour of treatment with exosomes (50 μg/mL), the conditional medium was replaced with fresh medium containing TGF-β at a final concentration of 5 ng/mL for 1 hour. Western blotting analysis revealed that p-Smad3 levels significantly increased after TGF-β (5 ng/mL) treatment (5.87-fold) compared to the control group (Figure 5A and B). Our findings suggest that liver cell damage induced by TGF-β can be ameliorated by WJ-MSCs exosome treatment, as evidenced by a significant downregulation of p-Smad3 levels to 2.86-fold (Figure 5A). Additionally, stimulation of the cells with LPS + Exo (50 μg/mL) led to an even further downregulation of p-Smad3, resulting in significantly lower levels than the Exo-treated group (1.94-fold).
The impact of 1-hour exosome treatment on transforming growth factor-beta (TGF-β)-induced Smad3 phosphorylation in the LX2 cell line was assessed. A, western blot gel image of phosphorylated Smad3 (p-Smad3) protein; B, p-Smad3 protein levels in LX2 cells treated with TGF-β, exosome and, exosome + lipopolysaccharide (LPS). The results, based on three replications (mean ± SEM), were reported, with a significance level set at P < 0.05. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) protein served as the internal control gene, and statistical significance was indicated as **** P < 0.0001, ## P < 0.01, ### P < 0.001.
5. Discussion
Liver injuries lead to wound formation, which acts as a primary driver of liver fibrosis. This process is facilitated by the activation of HSCs, crucial mesenchymal cells located in the liver's space of Disse. Pro-fibrogenic cytokines induce the proliferation and differentiation of HSCs into myofibroblast-like cells (25). Although HSCs are activated through various pro-fibrogenic pathways, TGF-β is one of the most prominent activators of ECM protein production by HSCs, as highlighted by multiple studies. Previous work has shown that miRNAs play major roles in regulating TGF-β signaling in HSCs (26). For instance, upregulation of miR-101 suppresses TGF-β signaling in HSCs by inhibiting TGF-βRI and KLF6 as its transcriptional activators (27). miR-19b inhibits TGF-β signaling by suppressing TGF-βRII and Smad3 gene expression. miR-378a targets TGF-β2, leading to inhibited expression of α-SMA and collagen I in TGF-β-treated HSCs (28).
Effective treatment of liver fibrosis may be achieved by inhibiting pro-fibrotic microRNAs. He et al. demonstrated that robust inhibition of miR-21 contributes to attenuating liver fibrosis in mice with hepatic schistosomiasis (29). Recently, miR-29b and miR-146a have gained attention as liver fibrosis inhibitors (30).
This study provides novel insights into the therapeutic potential of WJ-MSCs exosomes on liver fibrosis, focusing on modulating miR-29b, miR-146a, and the TGF-β/Smad3 signaling pathway. The exosome size distribution obtained from our zeta sizer readings is within the expected range for MSC-derived exosomes (31). The predominance of exosomes with a diameter of 73 nm is similar to findings by Cao et al., 2021, where MSC exosomes typically ranged from 50 - 200 nm (32). This uniformity in size distribution is critical for the reproducibility of therapeutic effects.
Our results show that TGF-β upregulates liver fibrosis markers, consistent with TGF-β's well-documented role in promoting fibrogenesis (7). Treatment with WJ-MSC exosomes resulted in a decline of these markers, suggesting an antifibrotic role of exosomes. This finding is consistent with recent studies, such as Salehipour Bavarsad et al., 2022, which demonstrated the potential of MSC-derived exosomes in mitigating fibrosis (33). The discrepancy in the effects of LPS + Exo treatment on α-SMA and collagen1α may indicate a complex interplay between exosomes and LPS, warranting further investigation.
The downregulation of miR-29b and miR-146a by TGF-β treatment adds to the body of evidence implicating these miRs in the fibrotic process (34). miR-29b targets multiple genes associated with ECM production (35), and its downregulation may contribute to fibrosis. Similarly, miR-146a has anti-inflammatory effects (36), and its downregulation could exacerbate fibrotic conditions. The significant upregulation of these miRs following Exo + LPS treatment highlights the possible modulatory role of LPS in exosome-mediated gene expression.
miR-29b has been extensively studied in the context of fibrotic diseases. It targets mRNA transcripts that code for ECM proteins such as collagen, fibrillin, and elastin (37). The downregulation of miR-29b observed after TGF-β treatment supports the hypothesis that reduced miR-29b levels may contribute to ECM component accumulation characteristic of liver fibrosis. This mechanism has been corroborated by other studies showing that miR-29b mimics can reduce collagen expression in fibrotic liver models (38), suggesting a therapeutic potential for miR-29b in attenuating fibrosis.
miR-146a is involved in regulating the innate immune response and inflammation. It negatively regulates NF-κB activation by targeting Tumor necrosis factor receptor associated factor 6 (TRAF6) and interleukin-1 receptor (IL1R)-associated kinase 1 (IRAK1), key adapter molecules in the toll-like receptor pathway (39). In liver fibrosis, inflammation is a critical driver of fibrogenesis, and downregulation of miR-146a could exacerbate this process by allowing uncontrolled NF-κB activity and subsequent pro-inflammatory cytokine production. Therefore, the upregulation of miR-146a by Exo + LPS treatment in our study might indicate a mechanism by which exosomes exert anti-inflammatory effects, potentially reducing the fibrogenic response.
The TGF-β/Smad3 pathway is central to the progression of liver fibrosis, with p-Smad3 acting as a key mediator (7). Our findings reveal that WJ-MSC exosome treatment can significantly downregulate p-Smad3 levels, suggesting an interruption of the TGF-β signaling pathway. This aligns with the work by Rashidi et al., 2023, where MSC-derived exosomes inhibited Smad3 phosphorylation (40). The further downregulation observed with LPS + Exo treatment suggests a potential synergistic or additive effect of LPS on exosome efficacy, which has not been extensively reported in the literature and opens new avenues for research.
5.1. Conclusions
Our study provides compelling evidence for the antifibrotic role of WJ-MSC-derived exosomes in the context of liver fibrosis. The size distribution of these exosomes aligns with the expected range, ensuring the reproducibility of therapeutic effects. Our findings corroborate the well-documented role of TGF-β in promoting fibrogenesis and reveal the potential of WJ-MSC exosomes in mitigating these effects.
The observed downregulation of microRNAs (miR-29b and miR-146a) by TGF-β treatment and their subsequent upregulation following Exo + LPS treatment underscores the complex interplay between exosomes and LPS. This highlights the potential modulatory role of LPS in exosome-mediated gene expression, opening new avenues for research.
Our study further elucidates the role of miR-29b and miR-146a in the fibrotic process. The downregulation of miR-29b, a known regulator of extracellular matrix production, and miR-146a, a key player in inflammation regulation, could contribute to the progression of fibrosis. The upregulation of these miRs following Exo + LPS treatment suggests a possible therapeutic potential for these miRs in attenuating fibrosis.
Finally, our findings reveal a significant downregulation of p-Smad3, a key mediator in the TGF-β/Smad3 pathway, following WJ-MSC exosome treatment. This suggests an interruption of the TGF-β signaling pathway, which is central to the progression of liver fibrosis. The further downregulation observed with LPS + Exo treatment indicates a potential synergistic or additive effect of LPS on exosome efficacy.
In summary, our study sheds light on the intricate mechanisms by which WJ-MSC exosomes and LPS may modulate gene expression and signaling pathways to exert antifibrotic effects. These findings pave the way for future research into the therapeutic potential of exosomes and specific miRs in the treatment of liver fibrosis.