1. Background
Hepatitis B virus (HBV) infection remains a significant global public health challenge, affecting approximately 260 million individuals worldwide who suffer from chronic infection (1). Annually, nearly one million people succumb to HBV-related ailments, including liver fibrosis, cirrhosis, and hepatocellular carcinoma (HCC) (1, 2). The development of liver fibrosis is a crucial factor in the prognosis of HBV-induced liver diseases. Strategies focused on eliminating agents responsible for triggering fibrotic responses may contribute to the regression of fibrosis (3, 4). Without intervention, HBV-associated liver fibrosis can advance to severe scarring and organ failure, exemplified by conditions like liver cirrhosis, ultimately progressing to HCC (3, 4).
The role of the immune response in controlling HBV is well established, particularly the contributions of adaptive immune responses involving virus-specific CD4+ and CD8+ T cells, B cells, and antibodies (5). However, the initiation of the innate immune response is paramount for achieving an adequate level of antiviral adaptive immunity. This initiation involves the recognition of conserved pathogen-associated molecular patterns (PAMPs) by cellular pattern recognition receptors (PRRs), marking a crucial initial step in mounting an effective antiviral defense (6). In the context of viral infections, nucleic acids act as conserved PAMPs, triggering the vigilant response of the innate immune system (6). Despite these advances, significant gaps remain in understanding the interplay between innate and adaptive immunity in HBV replication and control.
Nuclear DNA sensors are specialized proteins within the nucleus of cells that detect the presence of foreign or damaged DNA, triggering immune responses. In the context of viral infections, these sensors recognize viral DNA and activate signaling pathways, such as the production of interferons and other antiviral molecules to defend against the invading pathogen. Recent studies have highlighted the role of nuclear DNA sensors in antiviral host defense, including interferon (IFN)-inducible protein 16 (IFI16), cyclic GMP-AMP synthase (cGAS), IFIX, and heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNPA2B1) (7-11).
Interferon-inducible protein 16 has been recognized for its ability to associate with viral or transfected DNA, activate stimulator of interferon genes (STING), and coordinate IRF3 and NF-kB signaling in response to DNA viruses (11). Interferon-inducible protein 16's capacity to recognize viral DNA in both the cytoplasm and nucleus positions it as a versatile sentinel in the cellular defense against DNA viruses (12). The regulatory role of IFI16 is further complicated by post-translational modifications such as acetylation, which are integral to its function (13). The acetylation process is essential for the cytoplasmic translocation and subsequent signal transduction of IFI16 (13).
A novel aspect of this study is the investigation of the interaction between IFI16 and Sirt1, an NAD-dependent deacetylase known to reduce the acetylation of IFI16, thereby inhibiting its cytoplasmic localization and antiviral responses (14). Sirtuin 1, which has diverse regulatory roles in aging, metabolism, apoptosis, and inflammation, emerges as a potential partner in this molecular symphony (15). Recent investigations into Sirt1's role in antiviral responses, particularly in the context of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and HSV-1 infections, have revealed conflicting outcomes, highlighting the nuanced nature of Sirt1's involvement in viral replication (16).
Stimulator of IFN genes, which is downstream of IFI16, is an interferon-stimulated genes (ISG) and a key adapter protein in DNA-induced innate immune activation (17, 18). Given its crucial roles in activating innate immunity and autophagy, STING has emerged as a promising therapeutic target for a spectrum of diseases, including cancer, inflammatory conditions, and viral infections (19). Notably, agonist-induced STING signaling activation has been reported to boost antitumor immunity and may contribute to priming CD8+ T cells against immunogenic tumors, including HCC (20, 21). One study focuses on exploring the potential inhibitory effect of STING activation on HBV covalently closed circular DNA (cccDNA) and HBV-induced liver fibrosis, contributing valuable insights into the intricate interplay between STING signaling and the pathogenesis of HBV infection (22).
Our study aims to fill these gaps by investigating the collaborative efforts of IFI16 and Sirt1 in HBV replication. We focus on the molecular intricacies of their interaction and its impact on STING, a central player in regulating ISGs. Our exploration extends beyond the realm of IFI16 and Sirt1 to investigate the effects of their interaction on STING, shedding light on the regulatory network governing ISGs during HBV replication.
To further enrich our understanding of the host-virus interplay, we extend our investigation to the consequences of knocking down IFI16 and Sirt1. Surprisingly, the data reveal a scenario where knocking down IFI16 alone does not significantly affect HBV replication, but the dual inhibition of IFI16 and Sirt1 leads to a substantial reduction. As we navigate through the experimental landscape, our attention turns to the level of STING protein, a crucial component in the antiviral response. The observed changes in STING protein levels upon IFI16 and Sirt1 knockdown suggest a potential connection between IFI16, Sirt1, and STING in the regulation of ISGs and, consequently, HBV replication.
2. Objectives
This study sheds light on the intricate molecular ballet involving IFI16, Sirt1, STING, and ISGs in the context of HBV replication.
3. Methods
3.1. Vector Construction
A replication-competent 1.3mer, derived from the wild-type Hepatitis B Virus (HBV WT), was generously provided by Dr. Ryu WS at Yonsei University, South Korea.
3.2. Cell Culture and DNA Transfection
Huh7 cells are highly permissive to HBV infection, making them a suitable model for studying viral replication and host-cell interactions (23, 24). As a well-established hepatoma cell line extensively used in liver-related research, Huh7 cells provide a reliable and reproducible system for HBV studies (25-27). The cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 1% penicillin-streptomycin and 10% fetal bovine serum (FBS, Gibco BRL) at 37°C with 5% CO2. Passaging occurred every third day. For transfection into Huh7 cells, a mixture of 4 µg plasmid construct, 24 µg polyethylenimine (PEI, Polysciences), and 200 µl Opti-MEM (Gibco) was added to 1 × 106 Huh7 cells in 6 cm plates 24 hours after cell seeding. The cell culture medium containing transfected DNA was refreshed 24 hours post-transfection, and cells were harvested 72 hours post-transfection. "Mock-transfected" refers to a control condition in which cells undergo the transfection procedure without receiving the actual DNA or RNA of interest.
3.3. Core Particle Immunoblotting
At 72 hours post-transfection, cells were lysed using a 0.2% NP-40 (IGEPAL, Sigma-Aldrich)-TNE buffer [10 mM Tris-HCl (pH 8.0), 50 mM NaCl, 1 mM EDTA], following established protocols (25). Subsequently, 4% of the total lysate was electrophoresed in 1% native agarose gels. "Resolved core particles" refer to the HBV core particles that have been separated or "resolved" using native agarose gel electrophoresis. The resolved core particles were transferred to polyvinylidene fluoride (PVDF) membranes (Millipore). Immunoblotting to visualize core particles utilized a polyclonal rabbit anti-HBc primary antibody (1:1,000 dilution) (in-house generation), followed by a horseradish peroxidase-conjugated anti-rabbit secondary antibody (1 : 5,000 dilution) (Thermo Fisher Scientific). Bound secondary antibodies were visualized using enhanced chemiluminescence (ECL Western blotting detection reagent; Amersham). Relative core particle intensities were quantified using ImageJ 1.46r.
3.4. Nucleic Acid Blotting
To assess HBV DNA synthesis through Southern blotting, HBV DNA extracted from isolated core particles was separated on agarose gels, transferred to nylon membranes (Whatman #10416296), and hybridized with a 32P-labeled random-primed probe specific for the full-length HBV, following established protocols (25). Nucleic acid blotting was also performed on the same PVDF membranes used for core particle detection. Briefly, the PVDF membranes were treated with 0.2 N NaOH for 10 seconds, quickly washed with distilled water, dried, and then hybridized with a 32P-labeled random-primed probe specific for the full-length HBV sequence.
3.5. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis and Western Blotting
Equal quantities (determined via Bradford Assay) (28) of cell lysate [0.2% NP-40 (IGEPAL, Sigma-Aldrich)-TNE (10 mM Tris-HCl (pH 8.0), 50 mM NaCl, 1 mM EDTA) underwent sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 10% gels. "Resolved proteins" refers to proteins that have been separated by molecular weight through SDS-PAGE. The resolved proteins were transferred to PVDF membranes and incubated with appropriate primary antibodies: Rabbit polyclonal anti-HBc (1:1000), mouse monoclonal anti-IFI16 (1:1,000) (Santa Cruz #sc-8032), rabbit polyclonal anti-Sirt1 (1:1000) (Cell Signaling Technology #2493), rabbit monoclonal anti-STING (1:1000) (Cell Signaling Technology #13647), and mouse monoclonal anti-GAPDH (1:5000) (Santa Cruz #sc-32233). This was followed by incubation with anti-rabbit secondary antibodies coupled to horseradish peroxidase (1:5000 dilution) (Thermo Fisher Scientific) or anti-mouse secondary antibodies coupled to horseradish peroxidase (1:5000 dilution) (Thermo Fisher Scientific). Blots were visualized by ECL, and relative band intensities were measured using ImageJ 1.46r.
To ensure the quality and reproducibility of Western blot data, we followed standardized protocols and employed rigorous validation techniques. Specifically, protein samples were prepared using consistent methods across experiments, including appropriate lysis buffers and protein extraction procedures to maintain protein integrity. Additionally, loading controls were included on each gel to verify equal protein loading and transfer efficiency. Furthermore, to ensure the reproducibility of our results, Western blot experiments were independently replicated a minimum of three times under identical conditions. Quantification of band intensities was performed using ImageJ software, and statistical analyses were conducted to validate the significance of observed changes. These measures were implemented to uphold the quality and reproducibility of our Western blot data, thereby providing robust validation of protein-protein interactions and knockdown efficiencies.
3.6. Co-Immunoprecipitation
Huh7 cells were transfected with the 1.3mer HBV WT (ayw) construct, and harvesting occurred 3 days post-transfection. To determine the physical interaction between IFI16 and Sirt1, cell lysates were immunoprecipitated with rabbit polyclonal anti-Sirt1 and immunoblotted with mouse monoclonal anti-IFI16 antibodies. Rabbit normal IgG (Merck Millipore #12 - 370) served as a negative control for immunoprecipitation. Lysates underwent SDS-PAGE on 10% gels and were transferred to PVDF membranes for immunoblotting with primary antibodies (anti-IFI16, anti-GAPDH, and anti-Sirt1), followed by anti-mouse or anti-rabbit secondary antibodies coupled to horseradish peroxidase. Immunoblots were visualized using ECL (Western blotting detection reagent; Amersham). To validate protein-protein interactions, co-immunoprecipitation assays were performed using specific antibodies against the target proteins, with appropriate positive (indicating interactions) and negative (showing no interactions) controls included in each experiment.
3.7. RNA Interference
Sirtuin 1 siRNA (Santa Cruz #sc-40987), IFI16 siRNA (Santa Cruz #sc-35633), and negative-control siRNA (Invitrogen #4390843) were utilized. Huh7 cells were transfected with siRNA using Lipofectamine 2000 according to the manufacturer’s instructions. At 24 hours post-transfection, the cells were used for further experiments. To assess knockdown efficiencies, cells were transfected with specific siRNAs targeting the genes of interest, and knockdown efficiency was confirmed by Western blot analysis using validated antibodies.
3.8. Statistical Analysis
All experiments were performed three times, and images were captured using ImageJ. Data are presented as mean ± standard deviation and compared using Student’s t-test. P-values < 0.05 were considered statistically significant. Given the multiple experimental conditions tested, adjustments for multiple comparisons were made using the Bonferroni correction to ensure the validity of the statistical significance. This approach helps mitigate the risk of type I errors that can arise when conducting multiple statistical tests.
4. Results
4.1. Sirtuin 1 Interacts with Interferon-Inducible Protein 16, and This Interaction Enhances in Hepatitis B virus-Replicating Cells
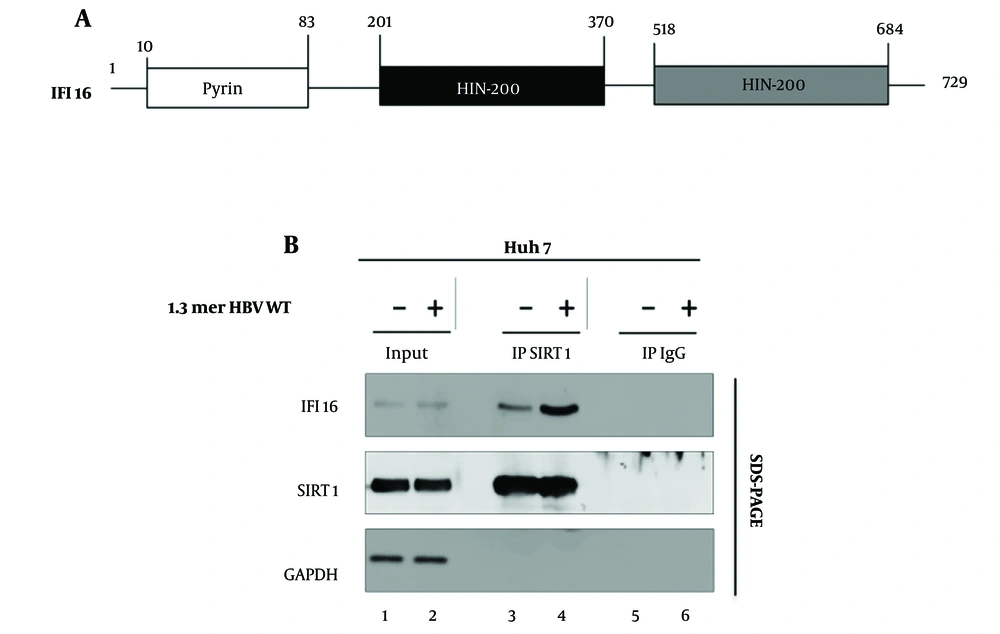
Previous studies have illuminated the multifaceted roles of Sirt1 and IFI16 in antiviral defense mechanisms, often linked to the induction of ISGs (14). Sirtuin 1, a NAD+-dependent deacetylase, has emerged as a key player in regulating diverse cellular processes, including DNA repair, inflammation, and stress response (15). Interferon-inducible protein 16, an IFN-inducible protein, has been recognized for its pivotal role in modulating the expression of ISGs, thereby contributing to innate immune responses, particularly in the detection of viral nucleic acids (11, 29) (Figure 1A).
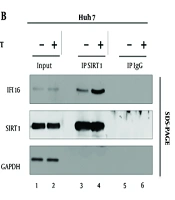
We turned our attention to HBV, a distinct viral entity with its own set of intricacies. Building upon the established Sirt1-IFI16 interaction in the context of HSV1, we sought to investigate whether a similar alliance existed in the realm of HBV replication. We examined the Sirt1-IFI16 interaction in HBV-replicating Huh7 cells (Figure 1B). Our data revealed an interaction between Sirt1 and IFI16 (Figure 1 lane 3) when HBV was not replicating in the cells. Astonishingly, our data revealed a dynamic shift, with the interaction intensifying significantly when HBV was actively replicating in Huh7 cells (Figure 1 lane 3 vs. 4). This finding hints at a novel layer of regulation specific to the interaction between Sirt1 and IFI16 during HBV infection.
Sirt1 interaction with IFI16 in Huh7 Cells. A, schematic of the full-length IFI16 protein structure; B, Hepatitis B virus (HBV) replication enhances the Sirt1/IFI16 interaction. Huh7 cells were either mock-transfected (lane 1) or transfected with 4 µg of wild-type Hepatitis B Virus (HBV WT) (lane 2). Lysates were subjected to immunoprecipitation using anti-Sirt1, followed by immunoblotting with anti-IFI16. Normal IgG served as a negative control. Sirt1 (Sirtuin 1), IFI16 (Interferon-Inducible Protein 16), GAPDH (loading control).
4.2. Interferon-Inducible Protein 16 and Sirtuin 1 Co-Knockdown Reduces Hepatitis B Virus Replication
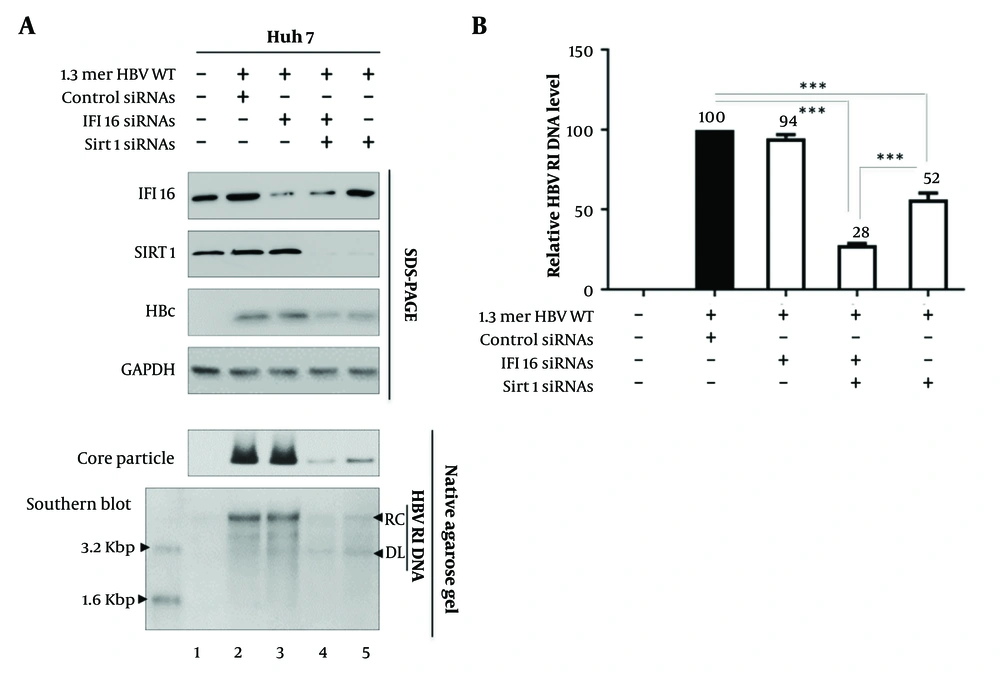
Since Sirt1 interacts with IFI16 and this interaction strengthens in HBV-replicating cells (Figure 1B), we next sought to unravel the distinct contributions of IFI16 and Sirt1 to HBV replication. We employed a targeted approach by utilizing siRNA to knock down these key players individually and in combination. The subsequent analysis of HBV replication dynamics provided novel insights into the specific roles of IFI16 and Sirt1 in this intricate process (Figure 2A).
Initially, we knocked down IFI16 using siRNA (Figure 2 lane 3). The data revealed that the depletion of IFI16 alone did not significantly impact HBV replication levels (Figure 2 lane 2 vs. 3). This observation challenges the conventional understanding of IFI16's role in antiviral responses, suggesting that, in the context of HBV, other factors may compensate for its absence or that IFI16 may function differently compared to its role in other viral infections. To explore potential synergistic effects, we extended our siRNA-mediated knockdown strategy to simultaneously target both IFI16 and Sirt1 (Figure 2 lane 4). Strikingly, our results demonstrated a notable reduction in HBV replication under these conditions (Figure 2 lanes 2 and 3 vs. 4). This finding suggests a cooperative relationship between IFI16 and Sirt1 in the context of HBV, implicating their concerted efforts in the regulation of viral replication. Intriguingly, when Sirt1 was targeted for knockdown alone (Figure 2 lane 5), the impact on HBV replication was not as pronounced as observed with the combined knockdown of IFI16 and Sirt1 (Figure 2 lane 4 vs. 5). Figure 2B presents a graphical representation of relative HBV DNA replication inhibition (RI) levels in the experimental settings explained in Figure 2A. Overall, this data shows that when Sirt1 and IFI16 are knocked down simultaneously, HBV replication is suppressed. This outcome underscores the specific role of Sirt1 in the regulation of HBV replication, offering a promising avenue for further exploration into the molecular intricacies of host-virus interactions in the context of HBV.
Sirtuin 1 (Sirt1) and interferon-inducible protein 16 (IFI16) co-knockdown reduces hepatitis B virus (HBV) Replication. A, Huh7 cells were transfected with wild-type Hepatitis B Virus (HBV WT), followed by treatment with control siRNAs (lane 2), IFI16 plus Sirt1 siRNAs (lane 4), or Sirt1 siRNAs alone (lane 5). Interferon-inducible protein 16, Sirt1, and HBc (Hepatitis B core) proteins were detected. GAPDH was used as a loading control. Core particles were analyzed by immunoblotting and Southern blotting to detect HBV replicative intermediate (RI), double-stranded linear (DL), and relaxed circular (RC) DNA. B, HBV RI DNA levels are shown as mean values from three independent experiments. HBV RI (replicative intermediate), DL (double-stranded linear), RC (relaxed circular). Statistical significance: P < 0.005 and P < 0.0005.
4.3. Interferon-Inducible Protein 16 Knockdown Reduces Stimulator of Interferon Genes Expression, but Sirtuin 1 Knockdown Enhances Stimulator of Interferon Genes Expression
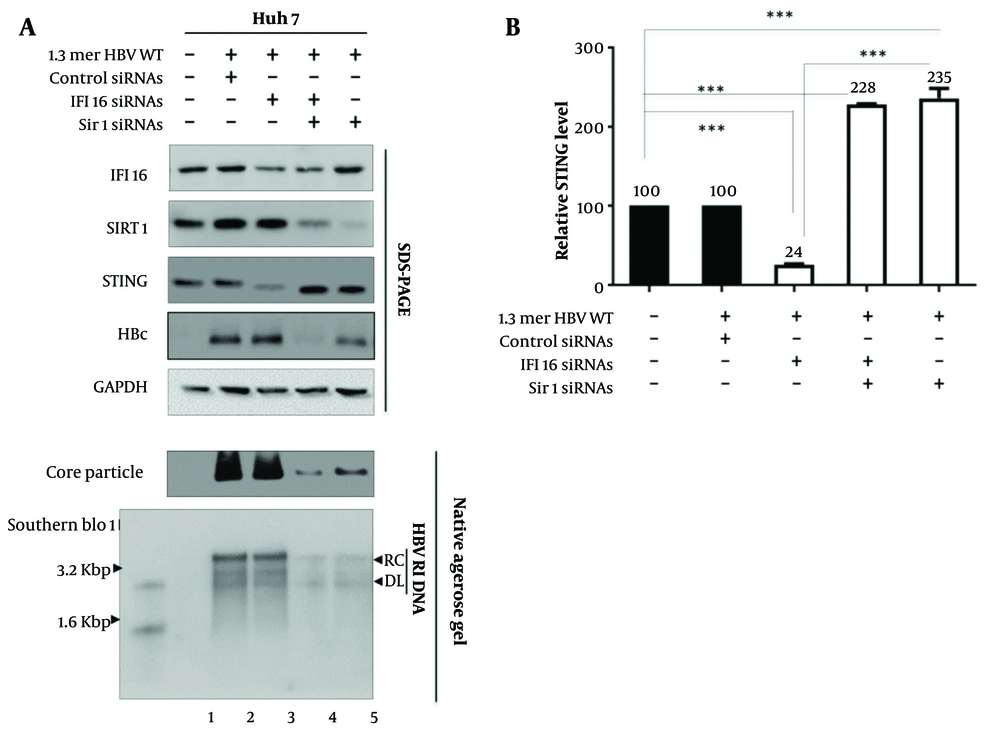
Building upon the foundation of our initial discoveries regarding IFI16 and Sirt1 in HBV replication, we delved deeper into the molecular intricacies by exploring the role of STING, a critical component in orchestrating innate immune responses. Stimulator of interferon genes, which acts downstream of IFI16, serves as a central hub in the detection of cytosolic DNA, including that from viral infections (17, 18). In the realm of host-virus interactions, numerous studies have established the crucial role of ISGs in shaping the cellular response to viral infections. Interferon-stimulated genes, including those regulated by the STING pathway, serve as molecular guardians that fortify cellular defenses against invading pathogens (17). Furthermore, it has been reported that Sirt1 prevents the interaction between STING and IFI16 during HSV-1 infection (14), prompting us to investigate the effects of Sirt1 on HBV replication (Figure 3A).
Upon knocking down IFI16, we observed a reduction in STING protein levels (Figure 3 lane 2 vs. 3). This finding suggests a potential regulatory connection between IFI16 and STING in the context of HBV infection. Given that STING is a key player in antiviral signaling pathways, the decrease in STING levels following IFI16 depletion may contribute to an altered cellular environment that facilitates HBV replication. Next, we performed a combined knockdown of IFI16 and Sirt1 (Figure 3 lane 4). While IFI16 knockdown alone reduced STING levels, the simultaneous depletion of Sirt1 and IFI16 resulted in an intriguing rebound of STING expression (Figure 3 lane 3 vs. 4). When Sirt1 was knocked down (Figure 3 lane 5), we noted an increase in STING protein levels (Figure 3 lane 2 vs. 5).
Sirtuin 1 (Sirt1) Knockdown Enhances stimulator of interferon genes (STING) Expression. A, Huh7 cells were transfected with wild-type Hepatitis B Virus (HBV WT) and treated with siRNAs targeting Sirt1 and interferon-inducible protein 16 (IFI16). Interferon-inducible protein 16, Sirt1, HBc, and STING proteins were visualized by immunoblotting; B, relative levels of STING protein were quantified using ImageJ. Data represent mean values from three independent experiments. Statistical significance: P < 0.005 and P < 0.0005.
This complex interplay between IFI16, Sirt1, and STING hints at a synergistic regulatory network where both proteins contribute to the modulation of STING levels in the context of HBV replication. In conclusion, our exploration of STING protein levels concerning IFI16 and Sirt1 modulation during HBV replication reveals a compelling link between these key players. The dynamic interplay observed underscores the complexity of the host response to viral challenges and sets the stage for further investigations into the molecular mechanisms that underpin the regulation of STING and its impact on antiviral defenses against HBV.
5. Discussion
Sirtuin 1, a versatile deacetylase with numerous substrates (30), plays a complex and controversial role in antiviral host defense. Its function depends on various conditions, such as host cell type, virus type, strain, and infection status. Recent evidence highlights Sirt1's involvement in HSV-1 infection, where Sirt1 negatively regulates HSV-1-induced antiviral innate immune responses (14). Sirtuin 1 serves as a negative regulator of DNA viruses or exogenous DNA-triggered innate immune responses in human cells. In our study, we examined Sirt1 as a regulator of HBV replication and delved into the dynamic interaction between Sirt1 and IFI16 in the context of HBV replication, shedding light on a novel alliance that intensifies during active viral replication.
Our investigation commenced by establishing the interaction between Sirt1 and IFI16 (Figure 1B), a phenomenon previously noted in the context of HSV-1 infections (14). Our data indicated that this interaction significantly intensified in HBV-replicating cells, suggesting a nuanced regulatory layer specific to the intricate relationship between Sirt1 and IFI16 during HBV infection. This novel finding prompted us to further dissect the individual contributions of Sirt1 and IFI16 to HBV replication.
Upon targeted knockdown experiments, we discovered that while the depletion of IFI16 alone did not markedly affect HBV replication, simultaneous knockdown of both IFI16 and Sirt1 led to a substantial reduction in viral replication (Figure 2A). This intriguing cooperative relationship between IFI16 and Sirt1 hinted at their concerted efforts in regulating HBV replication. Notably, the impact of Sirt1 knockdown alone was less pronounced, emphasizing the specific and collaborative role of Sirt1 in the complex interplay of host-virus interactions during HBV replication.
Our findings align with existing literature highlighting the multifaceted roles of Sirt1 in antiviral defense, particularly in HSV-1 infections, where Sirt1 negatively regulates antiviral innate immune responses by diminishing the acetylation of IFI16 and impeding its cytoplasmic distribution. While IFI16 is known to detect viral DNA in both the cytoplasm and nucleus, the recognition of herpesvirus genomes in the nucleus prompts IFI16 acetylation and subsequent cytoplasmic translocation. In the cytoplasm, IFI16 engages with STING, culminating in the phosphorylation of TBK1 and IRF3 and subsequent downstream signal transduction. Consequently, the cytoplasmic translocation of IFI16 following viral DNA sensing emerges as a critical step for its functionality in innate immune signal transduction. In HBV infection, a similar pathway may exist that warrants further exploration and experimentation.
Activation of STING, a central player in the regulation of ISGs, has proven to be a potent regulator that exerts a substantial inhibitory effect on cccDNA-driven transcription and HBV replication (31). Notably, this impact is more pronounced in the functional silencing of cccDNA rather than in altering the overall cccDNA levels. This study unveiled a fascinating aspect of STING signaling activation—its ability to induce heterochromatinization of cccDNA. This was evidenced by a simultaneous decrease in active chromatin markers, such as AcH3 and H3K4me3, and an increase in repressive chromatin markers, including H3K9me3 and H3K27me3, specifically on HBV cccDNA (31). Expanding our exploration, we also investigated the effects of STING on HBV replication. Interferon-inducible protein 16 knockdown resulted in reduced STING expression, suggesting a potential regulatory connection between IFI16 and STING in the context of HBV infection. However, simultaneous knockdown of Sirt1 and IFI16 led to a rebound in STING expression, revealing a complex interplay among these key players. Notably, Sirt1 knockdown alone enhanced STING expression, further underscoring the intricate regulatory network involving IFI16, Sirt1, and STING during HBV replication (Figure 3A).
In this research, we focused mainly on the effect of Sirt1 on IFI16-induced STING pathways in HBV infection. It would be meaningful to investigate the role of Sirt1 in IFI16-induced STING-TBK1-IRF3/NF-kB signaling pathways for HBV replication.
5.1. Conclusions
In conclusion, the correlation between IFI16, Sirt1, and STING hints at a potential regulatory network governing ISGs and, consequently, influencing the fate of HBV within the host cell. Sirt1 regulates the innate immune responses and ISG dynamics in HBV replication.
While this study provides valuable insights into the role of Sirt1 in regulating HBV replication through its interaction with IFI16 and the STING pathway, several limitations must be acknowledged. The experiments were primarily conducted using specific cell lines (Huh7), which may not fully replicate the in vivo environment of HBV infection in humans (32, 33). The responses observed in these cell lines might differ from those in primary human hepatocytes or in animal models. Additionally, for in vivo validation, animal models or clinical samples would be necessary to confirm the relevance of the findings in a physiological context.
The dynamic interaction between Sirt1 and IFI16 is critical during HBV replication. While knocking down IFI16 alone has minimal impact on HBV replication, the simultaneous knockdown of IFI16 and Sirt1 significantly reduces viral replication, suggesting a potential synergistic role. This research also explores the involvement of STING, a regulator of DNA-mediated innate immune activation, in the context of IFI16 and Sirt1 modulation during HBV replication. The findings indicate a complex interplay among IFI16, Sirt1, and STING, which may contribute to the regulatory network governing ISGs and innate immune responses. Overall, the study suggests that Sirt1 may play a specific and collaborative role in the intricate host-virus interactions during HBV replication, highlighting its potential as a promising target for further investigation and therapeutic interventions. Further studies are necessary to validate these interactions in vivo and to explore their therapeutic implications.