1. Background
Hepatic fibrosis results from abnormal tissue repair during wound healing, characterized by excessive deposition of hepatic extracellular matrix (1). Currently, there are only a limited number of effective antifibrotic drugs available for the clinical treatment of liver fibrosis (2). Metformin, a first-line drug for treating type 2 diabetes mellitus, has shown potential in addressing hepatic fibrosis. Metformin can alter intestinal flora and target hepatocytes to regulate protein synthesis, inhibit lipogenesis, and suppress gluconeogenesis (3). Consequently, it has been utilized in the treatment of hepatic fibrosis.
Metformin is known to inhibit the activation of hepatic stellate cells (HSCs), thereby reducing lipid accumulation, hepatic inflammation, and hepatic fibrosis (4). Additionally, probiotics have been employed in the treatment of liver fibrosis, achieving promising results (5). Metformin is absorbed into hepatocytes through Organic Cation Transporter 1 (OCT1) and has been shown to alter intestinal flora in both mice and humans. It has also been used to treat liver injury by improving sepsis-induced intestinal dysbiosis (6, 7). However, the mechanisms underlying its therapeutic action remain complex and are not yet fully understood.
The vast population of bacteria residing in the human gastrointestinal tract is collectively referred to as the human gut microbiome (8). These gut bacteria generally coexist harmoniously with their host, contributing to various physiological processes. However, dysbiosis of the gut microbiome can have detrimental effects on the host (9). Intestinal microorganisms influence liver function through the enterohepatic cycle, with endotoxins produced by harmful bacteria acting on the liver via this cycle, leading to liver damage (10).
Probiotic administration has been shown to reduce the incidence of hepatocellular carcinoma in mice, regulate the composition of intestinal microbiota, and restore intestinal permeability (11). For instance, the probiotic Lactobacillus rhamnosus GG has been demonstrated to ameliorate hepatic fibrosis by inhibiting bile acid synthesis (12). Additionally, the gut microbiota produces short-chain fatty acids (SCFAs), which play a crucial role in promoting intestinal barrier integrity (13). Conversely, dysbiosis triggered by a high-cholesterol diet has been linked to the progression of liver fibrosis and the development of hepatocellular carcinoma (14).
Environmental factors and pharmacological interventions, including drugs, can alter the composition of the gut microbiome.
2. Objectives
The primary aim of this study was to investigate the specific bacterial groups affected by metformin and to elucidate how these bacterial groups influence the progression of liver fibrosis.
3. Methods
3.1. Animal Models
Twenty-four C57BL6 male mice (3 - 4 weeks old) were obtained from the Animal Experimental Center of Guangxi Medical University. The mice were housed in a pathogen-free environment, provided with free access to food and water, and acclimatized for one week. Hepatic fibrosis was induced by intraperitoneal injection of a carbon tetrachloride (CCl4) solution in olive oil (Macklin, Shanghai, China) at a ratio of 1:4 (CCl4: Olive oil), administered at a dose of 1 mg/kg twice a week for four weeks. After the fibrosis induction phase, the mice were randomly divided into two groups (n = 12 per group): A control group, in which mice were treated with 0.9% NaCl (100 μL/day) for four weeks, and a metformin group, in which mice were treated with metformin (TCI, Japan, 200 mg/kg/day) for four weeks. At the end of the treatment cycle, feces and liver tissues were collected from all mice for further analysis.
3.2. Histology
Liver tissues were fixed, dehydrated, embedded, and cut into 3-μm thin sections. For morphological evaluation, the tissue sections were stained using the HE staining kit (Solarbio, Beijing, China) according to the manufacturer’s instructions. Sirius red staining (Saint-bio, Shanghai, China) was applied to assess the degree of liver fibrosis, and Oil Red O staining was used to evaluate the amount of lipid droplets in the liver samples, which were processed by Servicebio Technology Co., Ltd. All histopathological sections were imaged using a Nikon i80 microscope (Nikon Corporation, Japan). The positive areas for Sirius red and Oil Red O staining were quantified using Image J software (Image J 1.52a).
3.3. Immunohistochemistry
Tissue sections were subjected to antigen retrieval using sodium citrate antigen repair solution and blocked with endogenous peroxidase blocking buffer (Beyotime Biotechnology, Shanghai, China) for 10 minutes to inactivate endogenous peroxidase. The sections were then incubated overnight at 4°C with the primary antibody α-SMA (1:500, Servicebio, Wuhan, China) and subsequently incubated with horseradish peroxidase-labeled goat anti-rabbit antibody (1:50, Beyotime Biotechnology, Shanghai, China) at 37°C for 1 hour. Color development was achieved using diaminobenzidine (ZSGB-BIO, Beijing, China), and nuclei were counterstained with hematoxylin. All histopathological sections were imaged using a Nikon i80 microscope (Nikon Corporation, Japan). Quantitative immunohistochemical analysis was conducted using Image J software.
3.4. 16S RNA Sequencing Analysis of Mouse Feces
The collected mice fecal samples, stored at -80°C, were transported using dry ice to Shanghai Major Biological Company for DNA extraction and sequencing. Intestinal microbial DNA was extracted from the feces, and the 16S rRNA gene was amplified using PCR with TransStart FastPfu DNA polymerase (AP221-02) and the extracted DNA as a template. Barcoded primers 27F (5'-AGRGTTYGATYMTGGCTCAG-3') and 1492R (5'-RGYTACCTTGTTACGACTT-3') were used for the reaction. The PCR reaction system (20 μL) consisted of 5 × TransStart FastPfu buffer (4 μL), 2.5 mM dNTPs (2 μL), 5 µM upstream primer (0.8 μL), 5 µM downstream primer (0.8 μL), TransStart FastPfu DNA polymerase (0.4 μL), and 10 ng template DNA. The amplification procedure was carried out on an ABI GeneAmp® 9700 instrument with the following steps: Pre-denaturation at 95°C for 3 minutes, followed by 29 cycles of denaturation at 95°C for 30 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 45 seconds. This was followed by a final extension at 72°C for 10 minutes, and preservation at 4°C. Each sample was amplified in triplicate.
The PCR products from the same sample were combined and analyzed using 2% agarose gel electrophoresis. The bands were purified, and the products were quantified using a Quantus™ Fluorometer (Promega, USA). A library of the purified PCR products was constructed using the SMRTbell Barcoded Adapter Plate 3.0 (PacBio, USA). Library preparation included: (1) adaptor ligation; (2) magnetic bead screening to remove self-ligated fragments; (3) enrichment of library templates via PCR amplification; and (4) recovery of PCR products using magnetic beads to obtain the final library. Sequencing was performed on the PacBio Sequel IIe platform (PacBio, Shanghai Meiji Biomedical Technology Co., Ltd.) using the Sequel II Sequencing Kit 2.0 (1rxn) (PacBio, USA).
3.5. Statistical Analysis
Data analysis was conducted using GraphPad (version 8.0.2), and the results were expressed as mean ± standard deviation. Student's t-test was used to compare the means between the two groups. A P-value of < 0.05, < 0.01, < 0.001, or < 0.0001 was considered statistically significant.
4. Results
4.1. Metformin Attenuates Liver Fibrosis
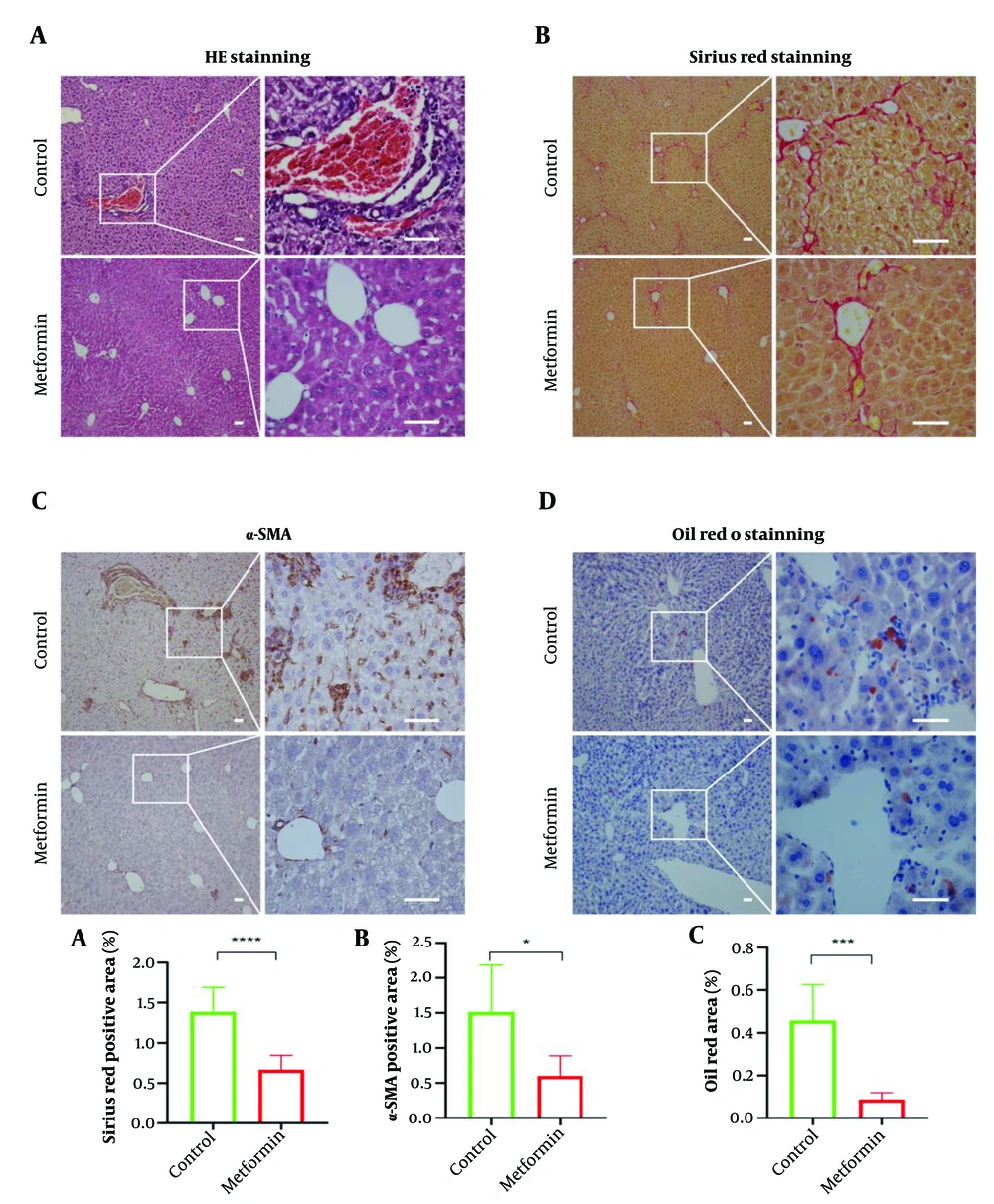
Liver fibrosis was induced in mice using CCl4. The mice were then treated with metformin (metformin group) or 0.9% NaCl (control group). At the end of the treatment period, liver tissues were collected for histological analysis. In the control group, hepatocytes exhibited edema and degeneration, with disorganized liver architecture, severe tissue damage, and extensive inflammatory cell infiltration. In contrast, the metformin-treated group displayed liver regeneration, reduced inflammatory cell infiltration, and an orderly liver structure (Figure 1A). Sirius red staining confirmed hepatic fibrosis and showed that metformin significantly attenuated fibrosis (P < 0.05, Figure 1B). In the CCl4-induced liver fibrosis model, α-SMA protein expression, a marker of hepatic stellate cell activation, was significantly elevated. However, α-SMA levels were significantly reduced in the metformin-treated group compared to the control group (Figure 1C). Furthermore, CCl4-induced liver fibrosis resulted in hepatic lipid deposition, which was significantly reduced by metformin treatment (P < 0.05, Figure 1D).
Metformin attenuates the degree of hepatic fibrosis and improves hepatic fat accumulation. (A), hematoxylin-eosin (HE) staining of liver (n = 6); (B), Sirius red staining of liver (n = 12); (C), immunohistochemical staining of liver with α-smooth muscle actin (α-SMA) (n = 6); and (D), oil red O staining of liver (n = 6). Scale bar: Fifty μm. Differences between the two groups were analyzed using an independent t-test, and data were expressed as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
4.2. Metformin Significantly Alters Intestinal Flora in Mice with Liver Fibrosis
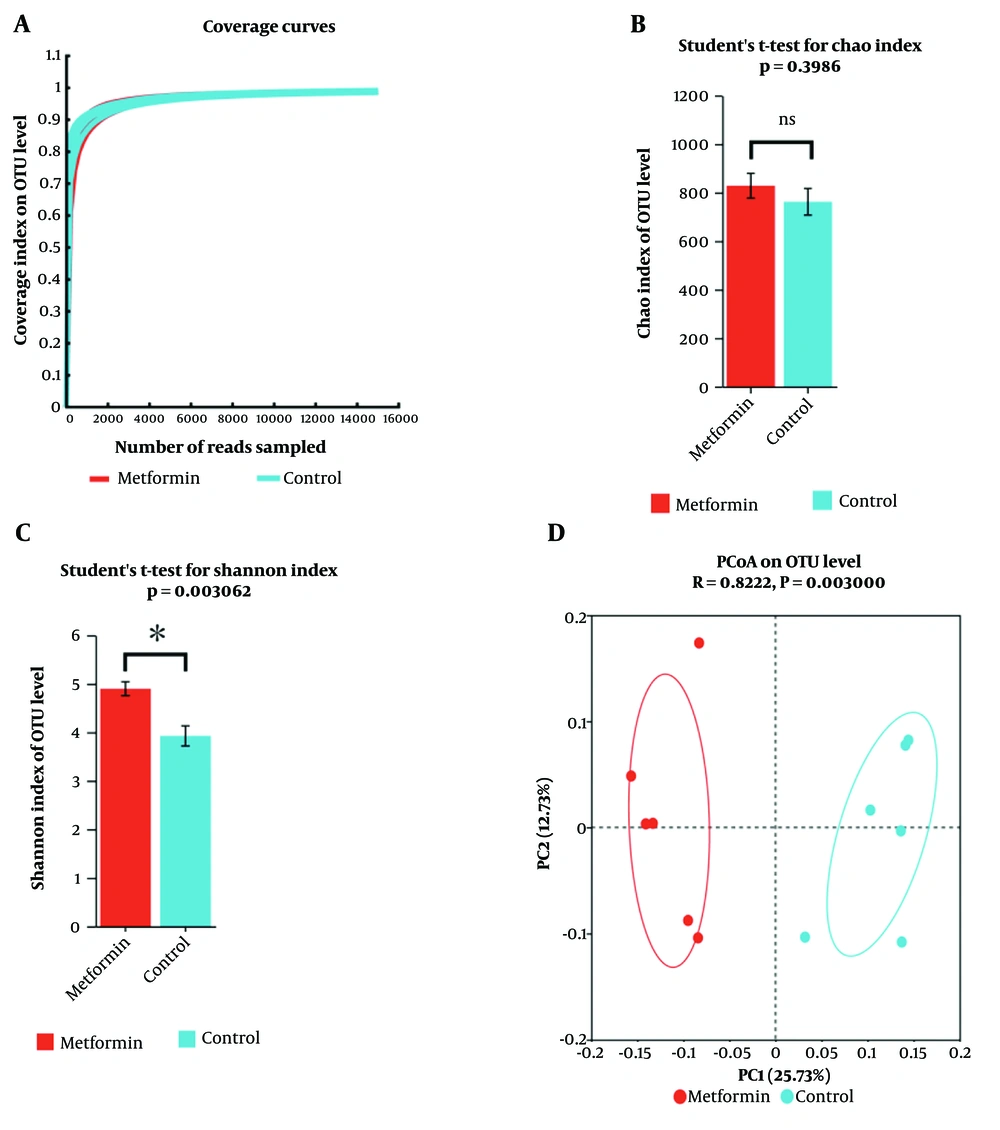
Gut analyses were conducted to evaluate whether metformin affects the gut microbiota. The dilution curve of gut flora (OTUs) demonstrated that the sequencing data reached saturation, indicating that it successfully captured the majority of species within the mouse gut microbiome community (Figure 2A). This confirms that the sequencing depth met the analytical requirements. Analysis of the Chao and Shannon indices (Figure 2B, C) revealed that metformin increased the species diversity of the mouse gut flora but did not alter its species richness. Principal coordinate analysis (PCoA) indicated significant differences in the gut microbiota composition of mice treated with metformin compared to those not treated with metformin.
α-diversity and β-diversity analysis of intestinal flora. (A) Coverage curve. (B) Chao Index analysis of treatment (metformin) and control groups. (C) Shannon Index analysis of treatment (metformin) and control groups. (D) Principal co-ordinate analysis of treatment (metformin) and control groups (Anoism, R = 0.82, P = 0.003, n = 6). * P < 0.05, ns indicates that the difference between the two groups is not statistically significant.
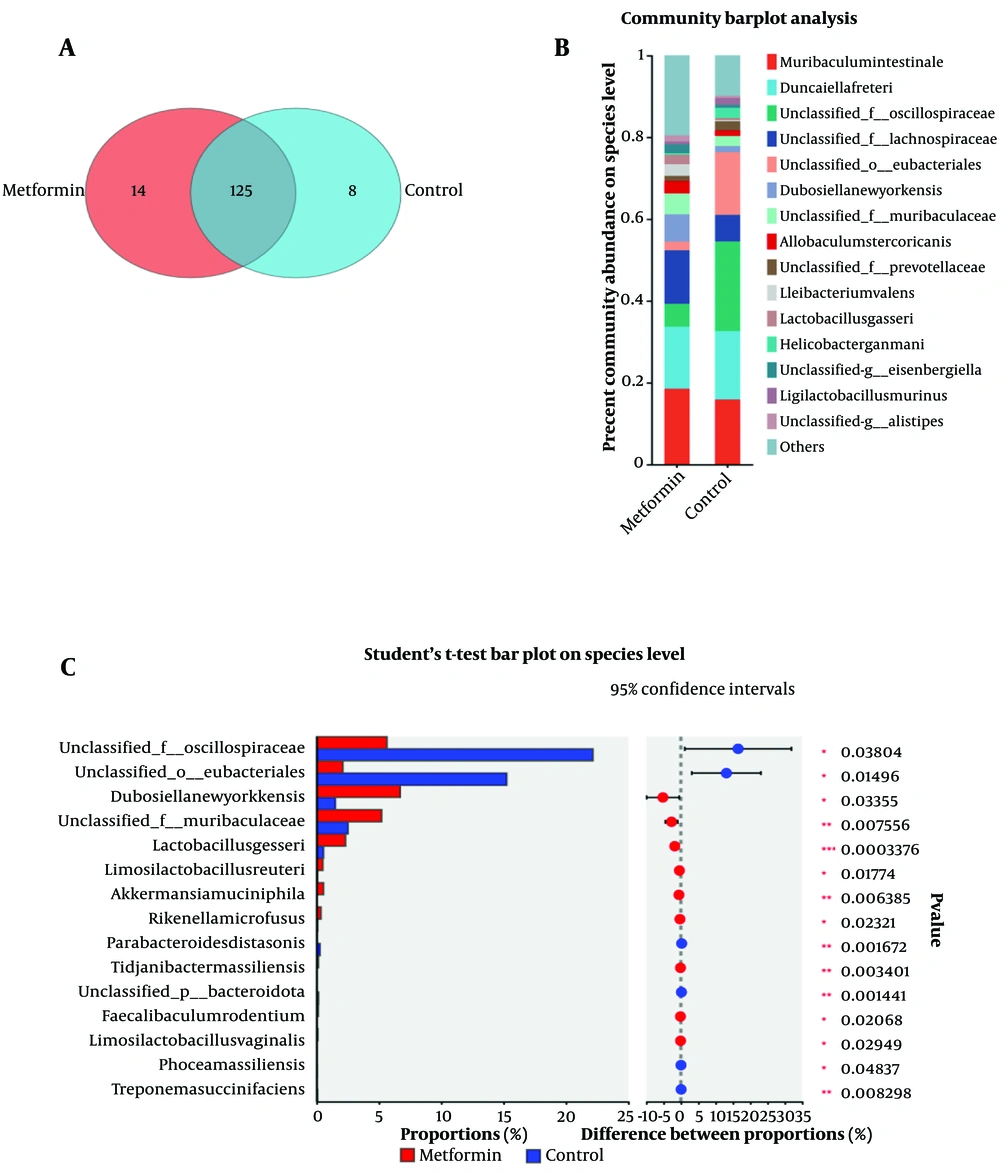
Venn diagrams showed that 125 bacterial species were shared between the metformin and control groups, 14 species were differentially abundant, and 9 species were unique to the control group (Figure 3A). Community bar plots identified the top 15 species with the highest abundance in each group (Figure 3B). Further analysis revealed 15 bacterial species with significant differences in abundance between the groups. Species with significantly higher populations in the metformin group, highlighted in red, included Dubosiellanewyorkensis, unclassified_f__Muribaculaceae, Lactobacillusgasseri, Limosilactobacillusreuteri, Akkermansiamuciniphila, Rikenellamicrofusus, Tidjanibactermassiliensis, Faecalibaculumrodentium, and Limosilactobacillusvaginalis. Species with significantly higher populations in the control group, shown in blue, included unclassified__f__Oscillospiraceae, unclassified__o__Eubacteriales, Parabacteroidesdistasonis, unclassified__p__Bacteroidota, Phoceamassiliensis, and Treponemasuccinifaciens (Figure 3C).
5. Discussion
Metformin treatment improves hepatic fibrosis; however, the underlying mechanism remains incompletely understood. This study aimed to investigate the effects of metformin on the gut microbiota. The abundance of Eubacteriales, Oscillospiraceae, and Helicobacterpylori was significantly higher in the control group. Notably, Oscillospiraceae abundance is markedly increased in non-alcoholic fatty liver disease (NAFLD) (15), suggesting that the elevated Oscillospiraceae in the gut of hepatic fibrosis mice may accelerate the progression of hepatic fibrosis.
In contrast, Faecalibaculumrodentium stabilizes the intestinal barrier, potentially preventing the release of bacterial endotoxins and protecting the liver (16). Helicobacterpylori, known to contribute to gastric cancer (17), has also been associated with an increased risk of developing NAFLD in infected individuals compared to non-infected individuals (18).
While Parabacteroidesdistasonis is known to provide metabolic benefits, such as improving insulin resistance and metabolic dysfunction (19), it was found in greater abundance in the control group than in the metformin-treated group. However, in both groups, Parabacteroidesdistasonis was present as a low-abundance bacterium, suggesting it may not have had a significant impact in either group.
There was a significant difference in the alpha and beta diversity of the gut flora between mice treated with metformin and untreated mice, suggesting that changes in gut microbiota are associated with metformin treatment. Metformin, a widely used drug for the treatment of type 2 diabetes, acts on the gut and induces alterations in gut flora (20). These metformin-induced changes in the gut microbiota play a beneficial role in many diseases and have garnered widespread attention (21). The liver and intestine are directly connected through the portal vein, allowing the transport of intestinal bacteria and the absorption of bacterial derivatives, which can significantly impact liver health (22).
For example, Lactobacillus rhamnosus GG has been shown to attenuate hepatic fibrosis by improving serum biochemistry, reducing inflammatory cytokine levels, and ameliorating histopathological damage in the liver and colon (12). Lactobacillus exhibits bile salt hydrolase activity, binding bile acids and increasing intestinal fibroblast growth factor 19 (FGF19) and hepatic small heterodimer partner (SHP) expression. This activity reduces cholesterol 7α-hydroxylase (CYP7A1) protein expression, subsequently decreasing bile acid synthesis, alleviating oxidative stress in piglets' livers, and preventing liver injury (23).
Dubosiella produces butyrate and SCFAs, which play a critical role in the progression of NAFLD and represent an important therapeutic target for NAFLD (24). Butyrate reduces hepatic steatosis by eliminating lipid deposits in the liver (25). In this study, metformin significantly reduced hepatic steatosis, likely due to its role in increasing Dubosiella abundance.
Additionally, Lachnospiraceae, a major producer of SCFAs, helps protect the intestinal barrier and inhibits the production of pro-inflammatory cytokines such as tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β), and interleukin-6 (IL-6). A reduction in Lachnospiraceae abundance has been implicated in pharmacological hepatic injuries (26). This study demonstrated that metformin significantly reduces liver inflammation and hepatic fat accumulation, further emphasizing its role in modulating gut microbiota and improving liver health. In summary, metformin increased the abundance of beneficial bacteria, decreased the prevalence of harmful bacteria, reduced liver inflammation, alleviated liver injury, and ultimately decreased liver fibrosis. This study, however, has some limitations. Due to various constraints, we were unable to determine the effects of metformin following the suppression of gut flora. Intestinal microorganisms represent a promising area for future research, given their significant connection to human health. Future studies should focus on exploring the intricate interactions between intestinal microorganisms and systemic health, particularly their role in liver diseases.
5.1. Conclusions
Metformin reduces hepatic fibrosis by modulating gut microbiota, thereby mitigating hepatic inflammation and lipid deposition.