1. Background
Up to 5% of the world’s population have been infected with hepatitis B virus (HBV), of whom probably 5% of the HBV carriers have hepatitis D virus (HDV) superinfection. It is estimated that 18 million people are infected with HDV world wild. Hepatitis D virus (HDV) is a defective single-stranded RNA virus which requires the HBsAg of HBV to establish infection in humans. The antigenomic strand of HDV encodes the only protein, hepatitis delta antigen (HDAg), in two molecular-weight forms. The large form carries an extra 19-amino-acid (aa) extension at the C terminus which plays a key role in the packaging of HDV and suppresses viral replication in a trans-dominant-negative manner, while the small HDAg plays an essential role in transactivating the replication of the HDV RNA (1). The disease spectrum of HDV infection varies greatly from fulminant hepatitis, rapidly progressive disease, to a subclinical course. Persistent replication of HDV associated with continuous hepatic inflammation and elevated alanine aminotransferase (ALT) levels is a characteristic of chronic active hepatitis D (2).
There are two modes of the HDV infection: coinfection, results from acute infection with both hepatitis B virus (HBV) and HDV, whereas superinfection results from HDV infection of patients with underlying chronic hepatitis B infection. Super infection with HDV increases higher risk of chronic HBV infection leads to progressive disease and cirrhosis in approximately 80% of cases than coinfection with HBV and HDV (3).
Based on phylogenic analysis, HDV isolates collected worldwide have been classified into 8 groups, HDV-1 (former genotype I), HDV-2 (former genotype IIa), HDV-3 (former genotype III), HDV-4 (former genotype IIb), and HDV-5 to HDV-8 (4). These HDV types show different geographic distribution and are associated with different disease patterns. Genotype 1 has been distributed worldwide, whereas other HDV genotypes circulate unevenly. The prevalence of HDV in Iranian hepatitis B infected individuals reported to be between 2.5% and 5.8% (5-7), and the major genotype was genotype I (8, 9).
2. Objectives
The aim of this study was to evaluate the prevalence of HDV in Iranian HBV isolates and to compare their clinical and virological pictures as well as their HDV genetic variations with other worldwide isolates.
3. Patients and Methods
3.1. Patients Design
81 patients with HBV infection, who were admitted to the Tehran Hepatitis Center (THC), Iran, for more than two years and who agreed to enter the investigation included in the study. Patients were chosen by the following criteria: positive results for HBs Ag, HBeAg negative chronic hepatitis or cirrhosis confirmed by histological and clinical evaluation, zero to low levels of HBV DNA, raised levels (5- 50 fold) of aspartate amino transferase (AST) and alanine amino transferase (ALT), no evidence of HIV or HCV coinfection, and no evidence of alcohol intake or exposure to hepatotoxic drugs. All patients had received IFN for two years. Informed consents were taken from patients.
3.2. RNA Extraction
Total RNA was extracted using acid guanidine phenol-chloroform method. In brief, 50µl of serum patient was mixed with lyses buffer (Qiagen), Proteinase K (Qiagen) RNA Carrier (Invitrogen) was incubated overnight. In aqueous phase, RNA was extracted by chloroform, and then the RNA was precipitated with isopropanol and washed twice with ethanol, dissolved in 200 µl of water, and stored at -80˚C until used.
3.3. Synthesis of cDNA for HDV RNA RT-PCR
9 µl of RT mixture, dNTP (10 mmol) (Roche), DTT (Roche, Germany), RT Enzyme (Invitrogen, Germany) and RNase (Promega, Germany) were transferred to 1.5 µl extracted RNA. Transcription was set up on ice and was performed as following thermal cycling: 25 ˚c for 5 minutes, 50 ˚c for 35 minutes, and 95 ˚c for 2 minutes. Subsequently, 10.5 µl of final cDNA solution was obtained from this reaction.
3.4. PCR Analysis of Hepatitis D Virus
Qualitative detection of HDV-RNA in serum was performed by single nested PCR amplification of a highly conserved region of HDV genome as described previously ( 10 ). For standard PCR, the region 1 (nucleotides 886 to 1288) corresponded to the C-terminal portion of the hepatitis delta Ag (HD Ag) coding region and included the RNA editing site and the poly adenylation signal (Smedile, et al. unpublished). The primers were designed according to an Italian HDV-infected genotype I (Table 1). Both single and nested PCR were performed by hot star Taq (Qiagen, Germany). The conditions of single PCR using primers EAB235 and EAB 237 set were: denaturation at 94˚C for 30 seconds, annealing at 64˚C for 1 min, and extension at 72˚C for 1 min for 40 cycles. The firs denaturation was performed at 94˚C for 2 min, and the last extension was performed at 72˚C for 10 min.
| Sequence | |
|---|---|
| First Round PCR | |
| PrimerEAB 235 HDV (858) | 5' GCC CAG GTC GGA CCG CGA GGA GGT 3’ |
| primerEAB 237 HDV (1312) | 5' ACA AGG AGA GGC AGG ATC ACC GAC 3’ |
| Second Round PCR | |
| primer EAB 236 HDV (883) | 5' GAA GGA AGG CCC TCG AGA ACA AGA 3’ |
| primer EAB 238 HDV (1288) | 5' GAG ATG CCA TGC CGA CCC GAA GAG 3’ |
3.5. Quantitative Determination of HDV-RNA and HBV-DNA
Serum HBV-DNA levels were determined by a commercial quantitative PCR assay (Amplicor HBV monitor test, Roche Diagnostics, GmbH Mannheim, Germany) with a sensitivity threshold of 2500 copies/ml (78UI/ml). HBV genotypes were determined based on reverse hybridization line probe assay.
3.6. Biochemistry and Serologic Tests
ALT, AST, alpha-fetoprotein, direct and total bilirubin, albumin, and prothrombin time were assessed by standard methods. Serologic markers of HBV, HAV and HCV were tested by commercial enzyme immunoassays (Abbot Laboratories, North Chicago, IL; Sorin Biomedica, Saluggia, Italy). Competitive enzyme immunoassay (ELISA) kit (DIA.PRO, Milano, Italy) was used for the detection of antibodies to hepatitis delta.
3.7. Sequencing
Amplified PCR products were purified by EXOSAP (USB Corporation, Cleveland, Ohio, USA) and obtained samples were directly sequenced by an automated sequencer (BMR-GENOMICS, Padova, Italy), using internal primers EAB 236 and EAB 238.
3.8. Mutational and Phylogenetic Analysis
Sequencing chromatograms were analyzed by Chromas (Chromas, version 2.3, Technelysium Pty Ltd, Tewantin, Australia) and Bioedit (5.0.9-tom Hall, Department of Microbiology, North Carolina state university) software. The alignment of HDAg amino acid sequences were performed by Clustal W (version 1.8-ftp.ebi.ac.uk). Final edited sequences were subjected for phylogenic analysis by applying Mega4 (version 4.1).
3.9. Indirect Immune Fluorescent Assay
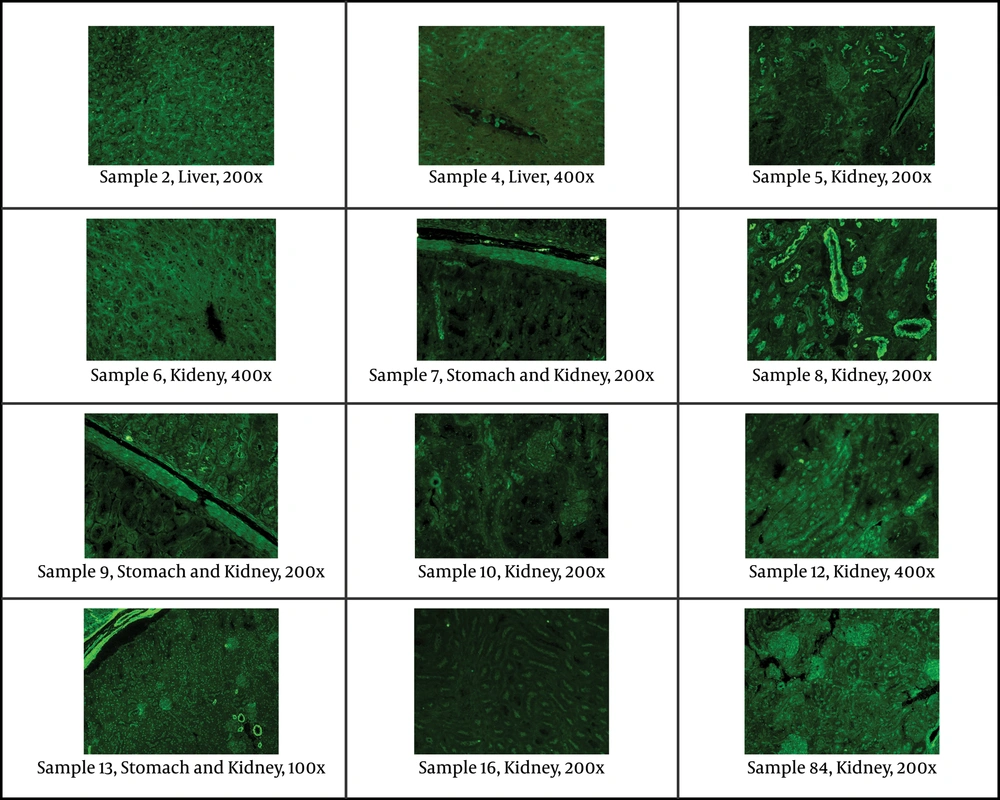
Indirect immunofluorescent assay was used on slides of mouse tissue (kidney, liver, stomach) to evaluate the main antibodies: ANA (anti-nuclear antibodies), ASMA (anti-smooth muscle antibodies), LKM (liver-kidney microsomes antibodies), ARA (anti-reticulin antibodies), AMA (anti-mitochondrial antibodies), anti-ribosomal antibodies, APCA (anti-parietal cell antibodies), and ABBA (anti-brush border antibodies). Serum samples were diluted 1:40 in PBS. A polyclonal rabbit anti-human IgA, IgG, IgM, Kappa, Lambda/FITC was used as fluorescent conjugate. On each slide tested, a negative control (IFA System Negative Control, NOVA Lite ANA KSL, INOVA Diagnostics), and a positive internal control (ANA homogenous pattern) were also tested in each essay.
4. Results
Of 81 individuals studied, 21 patients were female (25.9%) and 60 patients were male (74.1%), the mean age was 42. All of patients were of Iranian origin with different ethnic groups. Of the 81 serum samples tested with positive results for HBs-Ag, 12 (14.8%) patients had positive findings for both anti-HDV and HDV RNA (Table 2), of whom 5 and 7 were male and female, respectively. All had positive results for anti-HDV IgG as evidence of past infection, whereas, 8 had positive findings for anti-HDV IgM, suggestive of recent infection. The results revealed that all 12 isolates contained HBV genotype D and subgenotype D1. The details of patients with positive results for HDV-RNA are summarized in Table 2. All patients were HBs-Ag positive HBeAg negative with mean ALT and AST levels of 38.44 (IU/L) and 30.79 (IU/L), respectively. However, in patients who had positive results for HDV, the mean values for ALT level were 88.30 (IU/L) (kurtosis = 1.33) and 74.8 for AST (IU/L) (kurtosis = 1.33). All patients had a very low to moderate HBV DNA levels (results not shown); however, all HDV-positive samples showed very low levels (negative to 15 copy/ml) of HBV DNA (Table 2).
| Number | Sex | Age | ALT | AST | HBV Viral Load | HDV viral load, copy/ml | Anti-HDV IGM | Anti-HDV IGG | HDV- PCR | LIVER Biopsy | Number of Amino Acid Mutations | IF assay |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | f | 45 | 103 | 65 | < 15 | 1.45 E+08 145000000 | positive | positive | positive | cirrhosis | 12 | ANA+ 1:40 homogenous |
| 4 | f | 56 | 45 | 42 | NEGATIVE | 2.05 E+06 2050000 | negative | positive | positive | HCC | 11 | ANA+ 1:40 speckled |
| 5 | m | 50 | 87 | 83 | NEGATIVE | 1.47 E+06 1470000 | positive | positive | positive | chronic Hepatitis B | 15 | ABBA+ |
| 6 | f | 48 | 37 | 32 | 2673 | 3.62 E+07 36200000 | positive | positive | positive | cirrhosis | 8 | ANA+ > 1:40 speckled |
| 7 | f | 40 | 268 | 238 | < 15 | 3.84 E+07 38400000 | positive | positive | positive | Chronic Hepatitis B | 12 | ANA+ 1:80 homogenous |
| 8 | f | 58 | 90 | 73 | NEGATIVE | 2.17 E+08 217000000 | positive | positive | positive | 13 cirrhosis | 13 | ASMA+ > 1:80, ABBA+ |
| 9 | f | 58 | 38 | 30 | 240 | 5.28 E+05 528000 | negative | positive | positive | 16 cirrhosis | 15 | Negative |
| 10 | m | 62 | 90 | 113 | < 15 | 1.38 E+05 138000 | negative | positive | positive | cirrhosis | 11 | ANA + 1:80 homogenous |
| 12 | m | 53 | 24 | 47 | NEGATIVE | 1.29 E+06 1290000 | positive | positive | positive | chronic Hepatitis B | 22 | ANA + 1:40 fine speckled |
| 13 | m | 57 | 101 | 25 | NEGATIVE | 2.66 E+06 2660000 | positive | positive | positive | cirrhosis | 9 | ANA+ > 1:80 fine speckled, ASMA + > 1:80 |
| 16 | f | 53 | 43 | 54 | 344 | 1.44 E+05 144000 | negative | positive | positive | chronic Hepatitis B | 11 | ABBA+ |
| 84 | f | 49 | 35 | 42 | <15 | 8.69 E+08 869000000 | positive | positive | positive | Chronic Hepatitis B | 9 | ANA+ 1:80 homogenous |
4.1. Indirect Immunoflurescent
12 HDV-positive samples were studied by indirect immunoflurescent on mouse tissue for different main autoantibodies. All samples but one (sample No 9) had positive results for at least one autoantibody with different concentration and distribution patterns (Figure 1 and Table 2). Seven samples had positive results for ANA, and samples 8 and 13 had positive results for two autoantibodies for each (Figure 1 and Table 2).
Staining of Antinuclear Antibody (ANA), ABBA (anti-brush border antibody) and Smooth Muscle Antibody (ASMA) in Different Tissues of Patients With Positive Results for HDV Visualized by Indirect Immunofluorescence. The Concentration Cut off for Positivity of Antibodies as Well as the Magnification of Individual Samples Is Included.
4.2. Phylogenetic Analysis
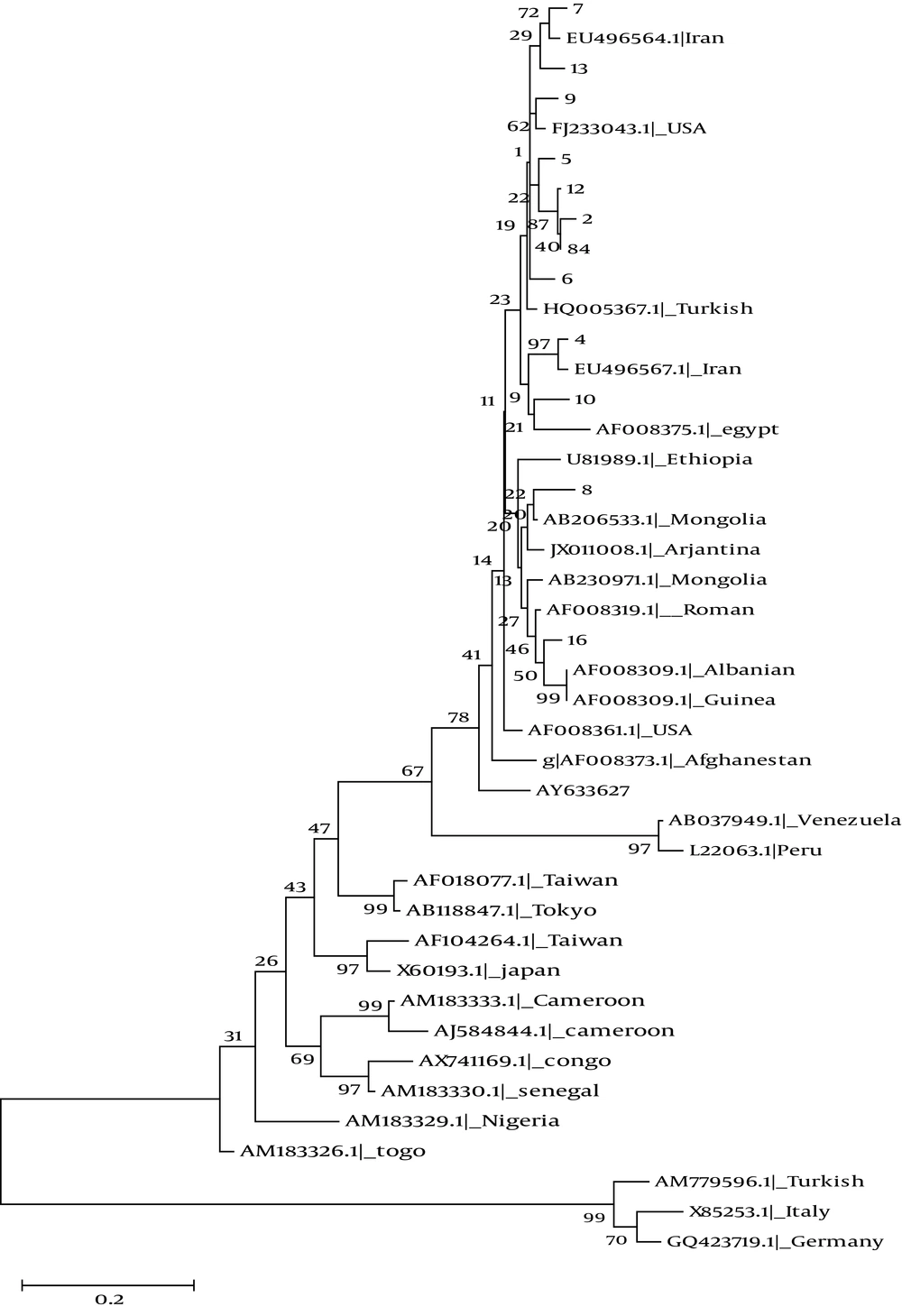
A 405- bp fragment of delta antigen taken from each of 12 patients with positive results for HDV-RNA was used for molecular study. The fragments were compared to the 29 reference gene sequences of HDV genotype 1 to 8 available in GenBank database (Figure 2). Genotype Ι was the only detected genotype. 10 of 12 isolates were branched into two separate clusters, and only two isolates (8 and 16) were grouped together with other genotype I sequences (Figure 2).
4.3. Mutation Analysis
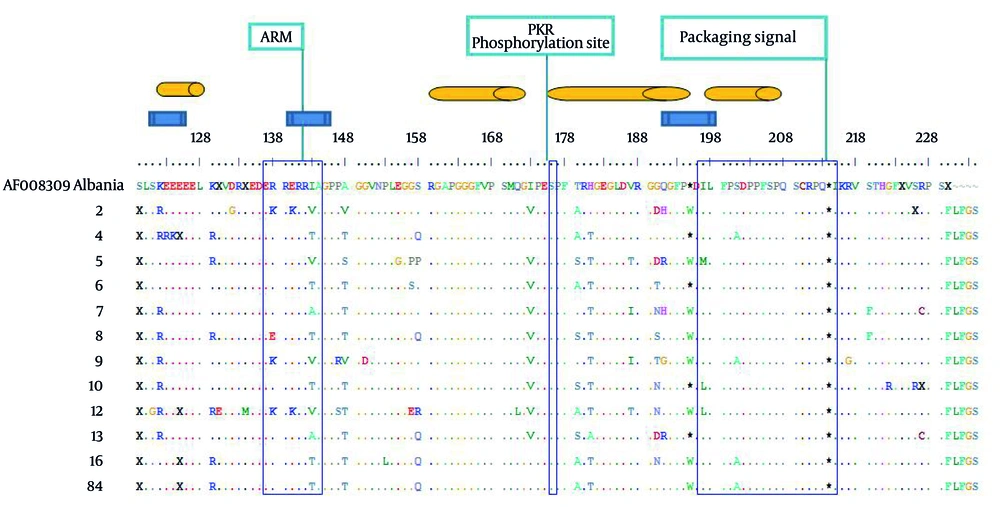
A genotype I isolate from Albania (Accession number AF008309) was chosen for sequencing alignment, due to the less average variations compared to other genotype I in the database sequences. There were 326 nucleotide mutations, of which 111 and 215 were silent and missense, respectively (results not shown). The ratio between silent and missense nucleotide mutations was 0.52. The total number of amino acid substitution was 148 (Figure 3). Within the region known for RNA binding domain (97-146), 18 amino acid mutations were occurred (Figure 3). Within virus assembly domain (195-204), four sequences contained stop codon mutations in position 195, on the other hand eight contained tryptophan (W), indicating expression of large delta antigens in the latter. Within the 19 C-terminus region of Delta Ag, seven point mutations were found (Figure 3). No mutation was found in PKR (Protein Kinase R) phosphorylation site (Figure 3).
72 mutations were distributed within four known different immune epitopes, of which 52 (72.2%) were occurred as a hotspot domain between amino acid residues 174 and 195 (Figure 3).
Note: The genotypic, functional, immune epitope domains are depicted in blue rectangle, open bars and orange cylinders, respectively. Variations along the partial small/large delta Ags were shown by letters in comparison to the reference sequence from a genotype I obtained from GenBank (accession number AF008309) from Albania. Dot indicates identity to the reference sequence.
4.4. Correlation Between Clinical and Virological Data
Overall, the mean ALT level was higher in patients with positive results for HDV (75.9 U/ML) than HBV-mono-infected individuals (results not shown). On the other hand, all HDV-positive cases but one showed a gradient decrease in the median HBV DNA load (from zero to 15 copy ml) compared to HBV mono-infected patients. However, there was no association between the grade of hepatic injury and HDV viral load. In mutational analysis, there were no significant correlations between the number and distribution of mutations and clinical, serological and virological details of patients (Table 2).
5. Discussion
In the present study, we evaluated the clinical and virological courses of HDV infection in a group of chronic HBV patients who showed a mild to moderate grades of hepatic injury, low to undetectable levels of HBV DNA, and a mild increase in the levels of liver enzymes. Biopsy-proven cases of cirrhosis and HCC also included in the patients-studied. While all the studied patients contained different low levels of HBV DNA, all HDV-positive cases but one showed a gradient decrease in the median HBV DNA load (from zero to 15 copy ml). This finding confirms the previous results (11) highlighting the suppression effect of HDV on HBV replication in these patients (12, 13). On the other hand, we could not find any association between HDV RNA load and the severity of liver disease including cirrhosis and HCC, the finding that was reported previously from Pakistan with the same distribution of HBV genotype D like Iran (14).
All samples but one (sample No 9) had positive results for at least one autoantibody with different concentration and distribution patterns. There are some reports in the literatures that HDV infection has been associated with autoimmune phenomenon (15, 16). However, the data that support this hypothesis is based on similar findings. The mechanisms and the significance of such autoantibody production have not been completely understood. Our result together with other results from Iranian studies showed that all HDV-positive cases had genotype I (9). Unpublished data from our lab indicated that all available Iranian HDV isolates have been closely related to European sequences. All these ethnic groups contained HBV genotype D dominantly, and all are of Caucasian origin. The distribution of HDV genotype I in Caucasian suggested that this ethnic group might have infected by the same HBV genotype (type D), then acquired HDV infection with the same genotype (type I) during colonization. Subsequently, due to intermixing between infected people and due to the random genetic drift, each virus undergone multiple variations. Previous finding might confirm this issue as data obtained from Turkey (17, 18), Lebanon (19), Russia (20), Mediterranean Basin and Europe (10, 21, 22), Mongolia (23, 24) and Tajikistan (25) which have shown the predominance of genotype I in genotype D infected HBV patients. The latter hypothesis is based on the finding of amino acid substitutions in residues involved in either immune epitopes and/or functional domains which are variants and not mutations. These positions also contained signature sequences regions specific for genotyping assignment (21, 26).
Patients with HBV/ HDV coinfection may have a more severe acute disease and in approximately 80% of these individuals the chronic HDV infection progresses to cirrhosis within 5-10 years (27). This observation is also noted in our HBV/HDV coinfection patients, as cases of HCC and cirrhosis were seen dominantly in HBV/HDV confection individuals as compared to HBV mono-infection cases. Worldwide, HDV genotype I has been associated with a highly pathogenic potential (28). This issue as well as the impact of HDV on the progression of liver disease in chronic HBV patients underscores the significance of diagnosis and genotyping of HDV in these patients to avoid such complications.
The ratio between silent and missense nucleotide mutations was 0.52, indicating a positive selection pressure exerted on these antigens. Among the four known B cell epitope residues (29) (Figure 3), 174-195 contained a hot spot motif for variation, as our results showed that it was covariated in all patients. Other epitopes showed only a few mutations in isolates. Due to the divergent structure of C-terminus domain of delta Ag (30), these variants seem to be the consequence of selection pressure imposed by host HLA during a long term process, which is different between versatile ethnic groups (31).
In the present study, the number of mutations in C-terminus part of Large-DAg (amino acid 195-214) was low (7 out of 148, 0.04%). Not surprising, as there are three conserved functional domains including nuclear exporting signal, clathrin heavy chain interacting domain and the isoprenylation signal in this short peptide region (32). Furthermore, several regions of RNA-binding sites and sequences in the glycine-prolyne rich segment (160-169, 175-179 and 183-187) are highly conserved among the HDV genotypes (17, 33). We found only two residues (179 and 187) in these domains which contained several mutations. Overall, of 10 residues which were contained mutations, all were contained in regions specific for genotypic classification and immune epitope function and replication (4, 34).
In conclusion, despite a high prevalence of 14.8% of HDV positive cases among chronic HBV carriers in our study, the limited number of samples evaluated and the cross-sectional study design prevented us to draw definitive conclusions. However, the initial suspension based on a florid HBsAg positivity and lack of HBV replicative markers underscore the importance of HDV diagnosis as an underlying disease which could accelerate progression of liver injury in these patients.