1. Background
Hepatitis C virus (HCV) is considered the most common cause of chronic liver diseases. It has been estimated that approximately 170-200 million individuals are infected with HCV all over the world (1) including about 17 million from Pakistan (2). HCV infection causes high morbidity and mortality rates due to development of cirrhosis and hepatocellular carcinoma and approximately 350000 people die every year globally due to HCV infection(3).
HCV genome is a 9.6 Kb single stranded RNA sequence flanked at both ends by untranslated regions (5 ‘UTR and 3' UTR) (4). A long single open reading frame is present in the coding region of HCV genome, which encodes a polyprotein of approximately 3000 amino acids. This polyprotein is processed posttranslationally by several viral and cellular proteins to produce at least 10 structural and nonstructural viral gene products (C, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B) (5, 6). HCV belongs to the genus hepacivirus in the Flaviviridae family classified into six major genotypes and multiple subtypes. These HCV genotypes and subtypes have variable distribution all over the world. Genotype 1a and 1b are the predominant genotypes in the United States and Europe, respectively (7, 8). HCV genotype 2 is particularly prevalent in countries of West Africa (4). Genotype 3a is more frequently found in Australia and South Asia (9), whereas genotype 4 is the predominant genotype in countries of Northern and Central Africa particularly Egypt. Moreover, genotypes 5 and 6 are frequently found in South Africa and Asia, respectively (4). All these HCV genotypes show 31-34% heterogeneity in their nucleotide sequences and approximately 30% heterogeneity in their amino acid sequences (10). Furthermore, HCV genotypes have their own unique pattern of disease development and response to antiviral therapy (11). To our knowledge, few studies have been conducted on distribution patterns of HCV genotypes in different geographical regions of Pakistan (12, 13). As the distribution of HCV genotypes can be changed over time, epidemiological studies may reveal distribution patterns of HCV genotypes among the Pakistani population to facilitate treatment options and preventive strategies.
2. Objectives
This study was performed to investigate distribution patterns of various HCV genotypes in different geographical regions of Pakistan.
3. Patients and Methods
3.1. Sampling
In this study, we evaluated patients with HCV infection primarily diagnosed with HCV infection by the third generation Enzyme Linked Immunosorbent Assay (ELISA) in local hospitals and then referred by physicians to the Chiragh Diagnostic Complex, Lahore for determination of HCV genotypes prior to antiviral therapy. Thousands of serum samples of patients were received at 30 collection centers of Chiragh Diagnostic Complex located at different districts of three provinces of Pakistan including Khyber Pakhtunkhwa, Punjab and Sindh between April 2011 and December 2013. From these a total of 2941 serum samples were randomly selected to detect and genotype HCV. The age of patients was between 16 and 56 years. Of these, 1573 were male and 1368 females. Consent was obtained from each patient for participation in this study at collection centers. Demographic information including gender, age and address were retrieved from patients' medical records.
3.2. Detection and Quantification of HCV RNA
HCV RNA was detected in serum samples of patients using FavorPrep TM viral RNA extraction Kit (FAVORGEN Biotech Corporation, ping-Tung 908, Taiwan) according to the manufacturer's instructions. Then for synthesis of complementary DNA (cDNA), 10 µL of HCV extracted RNA was incubated at 37°C for 50 minutes along with primers specific for core region and 200 U of Moloney Murine Leukemia Virus reverse transcriptase (MMLV RTase), (Thermo Fisher Scientific, Lenexa, KS 66285 USA), dNTPs and ddH2O. Then, cDNA was amplified using sense and antisense primers for qualitative analysis as described previously (14). PCR protocol was as follows: initial denaturation at 94°C for 5 minutes, followed by 45 cycles, each of 45 seconds denaturation at 92°C, 45 seconds annealing at 55°C and 1 minute extension at 72°C, with a final extension at 72°C for 10 minutes. Then, specific HCV PCR bands were visualized on 2% agarose gel stained with ethidium bromide in UV-transilluminator. HCV RNA was quantified in all PCR samples with positive results using PikoReal 24 Real time PCR system (ThermoFisher Scientific, Lenexa, KS 66285, USA) and AmpliSens HCV-FRT RNA quantification kit (Federal Budget Institute of Science, Moscow 111123, Russia) according to the manufacturer's instructions due to two kits. The lower detection limit of these assays was 5.0 × 102 IU/mL and its higher detection limit was 5.0 × 108 IU/mL.
3.3. HCV Genotyping
HCV genotyping was performed using type-specific HCV genotyping method as described previously (14). In brief, about 10 µL of HCV RNA was reverse transcribed to cDNA using 200 U of M-MLV RTase at 37°C for 50 minutes. Then, cDNA was amplified using primers specific for core region of HCV in the first round of PCR. Genotype with type specific primers for the core region of HCV genome was performed for the most common genotypes of HCV (1a, 1b, 1c, 2a, 2b, 3a, 3b, 4, 5a, and 6a). For discrimination of different HCV genotype amplified products, the type specific PCR mix was divided into mix A and mix B. Mix A contained primers for genotypes 1a, 1b, 1c, 3a, 3c and 4 and Mix-B included the 2a, 2b, 3b, 5a, and 6a primers. Obtained PCR products were then subjected to electrophoresis and separated on 2% agarose gel. After staining with ethidium bromide, gel was visualized under UV-transilluminator. HCV genotypes were determined by comparing the amplified product of a specific genotype with a 100-bp DNA ladder marker (Fermentas, USA), used as DNA size marker.
4. Results
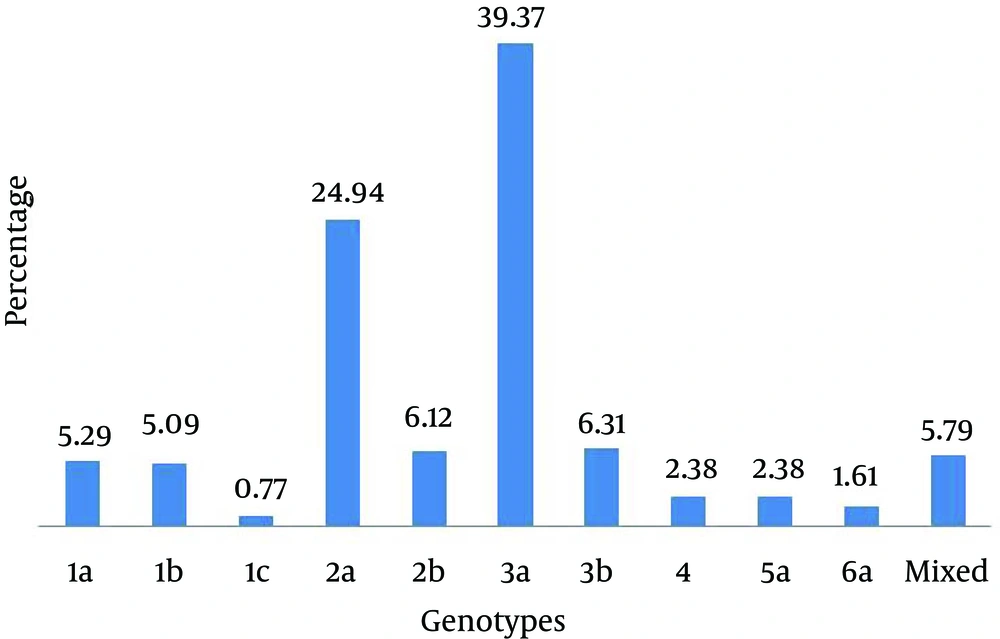
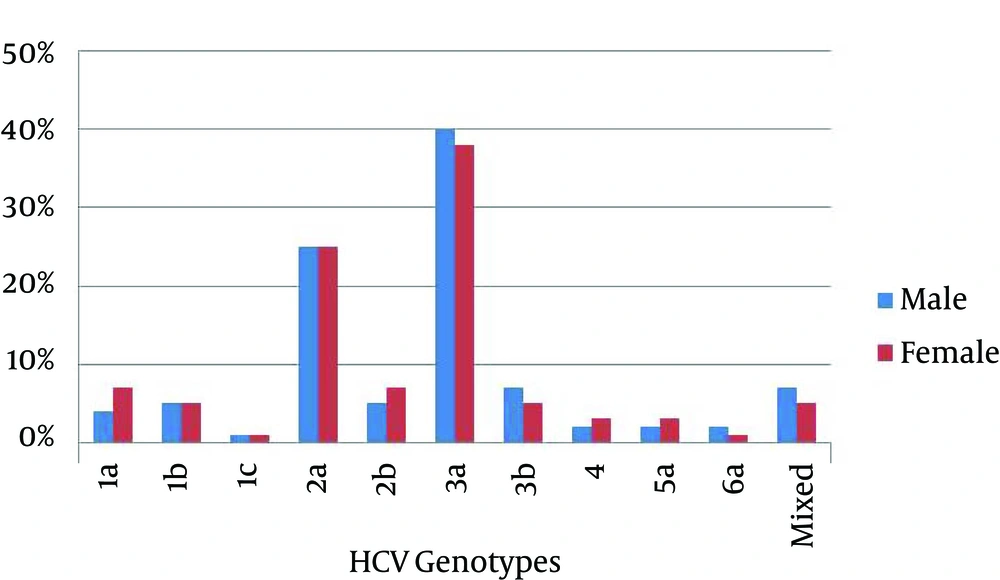
Of 2941 serum samples with positive results for anti-HCV, 1818 samples had positive findings for HCV RNA. After quantification of HCV RNA, 266 samples with low viral load < 500 IU/mL of HCV RNA were excluded from the study and 1552 samples with viral load > 500 IU/mL were analyzed by type-specific genotyping assay. In this study, 10 genotypes of HCV were identified as shown in Figure 1. HCV genotype 3a was the predominant one (39.4%), followed by genotypes 2a (24.93%), 3b (6.3%), 2b (6.1%), 1a (5.3%) and 1b (5.1%). HCV genotypes 4 and 5a were found equally distributed (2.38%) in studied patients. HCV genotypes 6a and 1c were found in only 1.6% and 0.7% patients, respectively. Moreover, 5.7% mixed genotypes of HCV were also detected in studied patients. Overall, the rate of HCV infection was comparatively higher in males (53.47%) than females (46.53%). Similarly, gender wise distribution of HCV genotypes was also determined among patients (Figure 2). Genotypes 3a and 2a were found most commonly distributed among both males and females patients. Genotypes 3a accounted for 337 (55.15%) male patients and 274 (44.85%) female patients; while, genotype 2a accounted for 212 (54.78%) male patients and 175 (45.22 %) female patients. In the current study, distribution of HCV genotypes in different geographical regions was investigated and presented in Table 1. Of 1552 patients, 728 (46.91%) were from Punjab, 541 (34.86%) from Khyber Pakhtunkhwa, and 283 (18.23%) from Sindh. HCV genotypes 3a and 3b were the predominant genotypes in Punjab region; while, genotypes 2a and 2b were the predominant ones in Khyber Pakhtunkhwa region. In Sindh region, genotype 3a was the most common genotype, followed by genotype 2a as the second common one. Moreover, genotype 1 was the predominant genotype in Punjab region compared to other geographical regions of Pakistan (Table 1). Distribution of HCV genotypes was also investigated according to different age groups (16-25 years, 26-35 years, 36-45 years, 46-55 years, and above 56 years) as presented in Table 2. Most patients (37.82%) were from the age group of 26-35 years, followed by 36-45 (34.09%), 16-25 (17.72%), 46-55(8.57%) and those above 56 years (1.80%). Therefore, higher incidence of various genotypes of HCV was observed among patients younger than 45 years compared to those older than 45 years as shown in Table 2.
| Geographical regions | HCV Genotypes | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1a | 1b | 1c | 2a | 2b | 3a | 3b | 4 | 5a | 6a | Mixed | Total | Age, y, % | |
| Khyber Pakhtunkhwa | 19 | 22 | 02 | 235 | 28 | 179 | 17 | 06 | 03 | 07 | 23 | 541 | 34.86 |
| Punjab | 45 | 38 | 07 | 114 | 38 | 336 | 47 | 21 | 23 | 09 | 50 | 728 | 46.91 |
| Sindh | 18 | 19 | 03 | 38 | 29 | 96 | 34 | 10 | 11 | 09 | 16 | 283 | 18.23 |
| Total | 82 | 79 | 12 | 387 | 95 | 611 | 98 | 37 | 37 | 25 | 89 | 1552 | 100 |
| Age groups | HCV Genotypes | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1a | 1b | 1c | 2a | 2b | 3a | 3b | 4 | 5a | 6a | Mixed | Total | Age, y, % | |
| 16-25 | 10 | 6 | 0 | 89 | 14 | 113 | 17 | 4 | 7 | 3 | 12 | 275 | 17.72 |
| 26-35 | 29 | 36 | 5 | 150 | 37 | 239 | 36 | 11 | 14 | 10 | 20 | 587 | 37.82 |
| 36-45 | 30 | 25 | 4 | 109 | 29 | 234 | 33 | 17 | 13 | 7 | 28 | 529 | 34.09 |
| 46-55 | 11 | 9 | 2 | 33 | 12 | 21 | 10 | 3 | 3 | 4 | 23 | 133 | 8.57 |
| 56-above | 2 | 3 | 1 | 6 | 3 | 4 | 2 | 2 | 0 | 1 | 6 | 28 | 1.80 |
| Total | 82 | 79 | 12 | 387 | 95 | 611 | 98 | 37 | 37 | 25 | 89 | 1552 | 100 |
5. Discussion
Studies on distribution of HCV genotypes have focused on mass screening of populations all over the world as they help to clarify the clinical status of HCV infection. Furthermore, they also facilitate treatment options and preventive strategies (15). Moreover, distribution patterns of Hepatitis C virus genotypes vary significantly in different countries and within the regions of the same country (16). Therefore, performing investigations on distribution patterns of HCV genotypes in our country is crucial for better understanding of HCV infection as well as implementation of preventive and therapeutic strategies. This study was conducted on 1818 patients with HCV infection from different geographical regions of Pakistan to evaluate distribution patterns of HCV genotypes within this population. The present study is in concordance with previous studies, in which HCV genotypes were investigated from different regions of the world (16-18). Predominance of genotype 3a was found in this study and previous studies from Pakistan (11, 12, 19). Genotype 3a has also been the predominant genotype in Afghanistan and India (20, 21). Moreover, genotypes 1a and 1bwere common in Iran and China, respectively (18, 22). The results of this study were comparable with a previous study on HCV genotypes distribution in Pakistan (12), it was found that the incidence of genotype 2 is increasing in Pakistan with decrease in the incidence of genotype 3. This corresponds well to the previous study that in next 15-20 years, the most prevalent genotype 3 would be replaced by less prevalent genotypes in Pakistan. The overall incidence of genotype 1 was found similar in both studies, but we suggest further studies to confirm these observations. Studies from Germany and Venezuela reported replacement of HCV predominant genotypes with less prevalent ones (23, 24). To determine regional differences in HCV genotypes distribution in Pakistan, HCV infected patients from three provinces were included. An obvious difference was found in Khyber Pakhtunkhwa province, where the genotype 2a was found the predominant genotype followed by 3a (Table 1). This is consistent with the result of a previous study from Pakistan where genotype 2a was the predominant genotype in IDUs in Khyber Pakhtunkhwa (25). However, no regional difference was observed regarding genotypes distribution of HCV in Punjab and Sindh provinces, where the predominant genotype was 3a followed by 3b (Table 1). Gender wise distribution of HCV genotypes was also observed in this study. Distribution of HCV genotypes was found variable among male and female patients. Interestingly, it was observed that the predominant genotypes in males were 1b, 1c, 3a, 3b, 5a and 6a versus 1a, 2a, 2b and 4 in females (Figure 2). In contrast to our observation, no gender wise variation was reported in HCV genotypes distribution in Algeria (17). However, in Libya, HCV genotype 1 was frequently found in males, whereas genotype 4 was frequently found in females (16). Many studies have suggested that distribution of HCV genotypes may be inconsistent among patients of different age groups. Previous studies reported that genotype 1a was found more commonly in younger individuals, while genotypes 1b, 2a and 2b were more commonly found in older patients (26, 27). In Iran, genotype 3a was the predominant genotype in patients younger than 40 years (18). Another study from France reported that genotype 5 was the commonly found genotype in older patients (28). In this study, we observed distribution of HCV genotypes among different age groups (Table 2). Incidence of HCV genotypes, particularly 3a and 2a was high among patients younger than 45 years compared to older ones (> 45 years). Further studies are needed to determine possible risk factors in transmission of HCV for effective implementation of preventive strategies to reduce young people exposure to this infection.
In conclusion, we observed that the HCV genotype 3a is the predominant genotype in Pakistan followed by 2a. The incidence of genotype 2a was observed to be increasing in our country with decrease in the incidence of genotype 3a. Regarding regional differences in Pakistan, genotype 2a in the Khyber Pakhtunkhwa province and genotype 3a in the Punjab and Sindh provinces were the predominant genotypes. HCV genotypes distribution was found variable based on age and gender. Periodically investigations of HCV genotypes are needed to monitor distribution patterns of genotypes to facilitate treatment options and preventive strategies in our country Pakistan.