1. Context
Hepatitis C virus (HCV) is a global public health problem. About 170 million people, nearly 3% of the world’s population, are chronically infected and at risk of developing liver diseases including cirrhosis and hepatocellular carcinoma. It has been estimated that three to four million are newly infected and 350000 deaths occur each year due to all HCV-related diseases. Liver dysfunction by HCV infection is one indication for liver transplantation. No vaccine or immune-globulins are available for prevention. Low effectiveness and high cost of the current antiviral drugs limit the treatments of HCV infection (1, 2).
Nowadays the most common route of HCV transmission is through syringe sharing among intravenous drug users (IVDUs) with a prevalence rate of 31 to 89% in different geographical areas of the world. Risk of infection in some groups such as, patients on hemodialysis, hemophilia, infants of mothers with HCV infection, and multi-partner individuals is very high. Prevalence of HCV among patients on hemodialysis was 5.5% to 55.9% in different cities of Iran, although this number for patients with hemophilia was 15.65 to 76.7% (3). The rate of HCV positivity among IVDUs was 52% in Tehran (4).
According to a recently conducted meta-analysis, the overall prevalence of HCV among blood donors in Iran’s provinces and cities was 0.5% (95% CI: 0.4-0.6%). The highest prevalence of HCV was in Kashan (1.09%), and the lowest prevalence was in South Khorasan (0.03%). Hepatitis C virus prevalence had a decreasing trend over 11 years. The highest prevalence rate was 1.39% in 2005; then, the prevalence rate had a decreasing trend to 0.13% in 2007 and 0.03% in 2009 (5).
In Iran HCV prevalence in the general population is significantly lower when compared to the corresponding populations in surrounding countries such as Afghanistan (1.1%), Pakistan (4.7%) (6), Turkey (1.0-2.1%), Kuwait (0.8%) and Iraq (7.1%) (7).
Hepatitis C virus is a small, enveloped virus that belongs to the genus Hepacivirus and the family Flaviviridae. The diameter of the virion is about 55-65 nm. The genome of HCV is a positive single-stranded RNA molecule of approximately 9.6 kb. It encodes a polyprotein from a single open reading frame flanked by two untranslated regions (UTR) at the 5' and 3' ends of the genome. This polyprotein precursor is cleaved by proteases giving rise to structural (Core, glycoproteins E1, E2) and nonstructural proteins (NS2, NS3, NS4A, NS4B, NS5A and NS5B). The HCV genome has a high rate of mutations therefore genetic heterogeneity of HCV is observed in patients (2, 8).
Hepatitis C virus is classified into seven major genotypes (1, 2, 3, 4, 5, 6 and 7) and more than 67 subtypes (named a, b, c, etc.) (9). Sequence differences range from 31% to 33% among genotypes and from 20% to 25% among subtypes. Distribution of HCV genotypes is variable. Genotypes 1 to 3 have a worldwide prevalence, whereas genotypes 4 to 6 are more geographically restricted. Genotype 4 is found mainly in North Africa and the Middle East, especially in Egypt. Genotype 5 is restricted to South Africa and genotype 6 is found in South East Asia (10). The most dominant subtypes in south and north-west of Iran are 1a and 3a, respectively, while 1a predominates in all parts of Iran (11, 12).
Hepatitis C virus genotyping is important for the management of infected individuals and for epidemiology purposes (10). Genotypes indicate the route of acquisition, and affect the clinical outcome and response to treatment. For example response to interferon therapies in combination with ribavirin in patients infected with genotypes 1 and 4 is much lower than genotypes 2 and 3. Infection with genotype 1 may proceed rapidly to severe chronic disease, cirrhosis, and hepatocellular carcinoma. Studies have shown that subtype 3a is significantly associated with transmission through IVDUs in industrial countries (4). Therefore, before any treatment, the genotype of the virus must be determined to estimate the dose, duration and type of antiviral drugs, and the virological monitoring method (10, 12).
2. Objectives
There are studies on the prevalence of HCV genotypes in infected individuals from Iran, yet they have not provided information about the overall distribution of HCV genotypes. Knowledge of the prevalence and distribution of HCV genotypes in infected individuals is important for successful treatment, control strategies and researches in the future. The aim of this study was to estimate the overall prevalence and trend of HCV genotypes in Iranian patients.
3. Data Sources
3.1. Search Strategy and Selection Criteria
The literature was searched to find scientific papers reporting the prevalence and distribution of HCV genotypes in Iran. Papers were identified through searching PubMed, Magiran, IranMedex, Scientific Information Databank (SID), and Google scholar databases. To search for as many related studies as possible, we used keywords including: “hepatitis C virus or HCV”, “genotypes”, “molecular epidemiology” and “Iran”. We also manually searched relevant Persian journals.
4. Study Selection
The inclusion criteria were:
full text articles in English or Persian language reporting on HCV genotypes among infected patients with Iranian nationality;
studies using HCV RNA as a detection index of infected subjects that had data indicating the prevalence of HCV genotypes using standard genotyping methods.
The exclusion criteria were:
studies with unknown sample origins;
studies with overlapping subjects, time and place of sample collection;
studies that failed to present data clearly;
any contraries to the inclusion criteria.
5. Data Extraction
All potentially relevant papers were reviewed independently by two investigators that assessed the eligibility of each article and the data were abstracted by the Excel data sheet. Irrelevant papers and papers that did not clearly report HCV genotypes were excluded. The following information were extracted from the papers: first author’s name, study period (when the study was conducted), province or city of sampling, patients population, sample size, HCV genotypes and subtypes, genotype detection methods, gender, age, risk factors, etc.; although some papers did not contain all of these information.
For studies with duration of more than one year, the middle period of the time frame was used for analysis of trend. In addition, for some studies in which the prevalence of genotypes was reported annually, prevalence results of each year was considered in trend analysis. Our analysis was in accordance with the items of systematic reviews and meta-analyses (PRISMA) guidelines for systematic review.
5.1. Statistical Analysis
We considered studies reporting HCV genotypes (or subtypes) in Iran and 95% confidence interval were calculated using random effects model and meta-analysis. The possibility of significant heterogeneity between studies was tested with the Q test (P < 0.10 indicated statistically significant heterogeneity) and the I2 statistic. Freeman-Tukey’s double arcsine transformation was used to stabilize variances. In addition, Forest plot was performed for study locations and years. Data manipulation and statistical analyzes were done using the Meta R statistical software package, version 2.13. In meta-analyses with sufficient number of published papers, a funnel plot was inspected visually to assess for publication bias.
6. Results
6.1. General Information of Samples
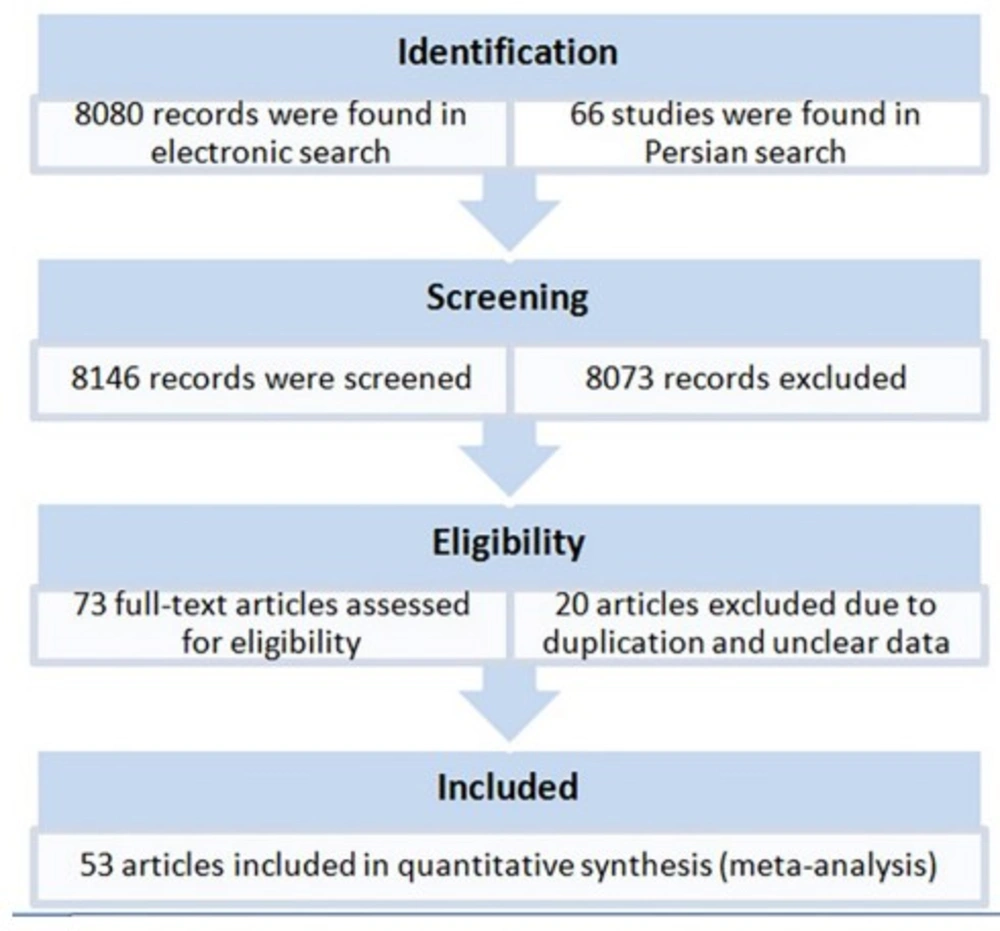
A total of 73 original articles were found by the electronic search that reported HCV genotypes and subtypes from provinces and cities of Iran (Figure 1). After exclusion of ineligible papers, the final sample of the meta-analysis included 53 studies published between 1999 and 31 June 2014, including 44 studies in English, and 12 studies in Persian. Studies were done in 20 provinces or cities of Iran. In total, 22952 patients were meta-analyzed for pooled prevalence of HCV genotypes (Table 1).
The populations of studies were mainly chronic HCV infected patients including patients with hemophilia and thalassemia as well as those on hemodialysis, and multiply transfusion. In addition, a few studies included patients with a history of prison, dental visits, tattooing, unprotected sexual contact, kidney transplantation, intravenous drug use, and blood donation (Table 1). Since the genotypes and subtypes in the patients' populations were not reported separately, we were unable to analyze the prevalence of genotypes and subtypes according to population groups. Gender of patients was mostly male (45% to 100%) (Table 1). As genotypes according to patients' gender were not reported by all papers, we were unable to analyze the prevalence of genotypes according to gender. Due to data limitation, subtypes 2a and 2b were not analyzed. Data regarding risk factors were not complete in published papers thus their analysis was not possible.
Different methods have been used for HCV genotyping in Iranian patients, including 5´-UTR restriction fragment length polymorphism (RFLP), DNA sequencing of NS5B or 5'-UTR/core genes, high resolution melting analysis, nucleic acid hybridization, and type-specific PCR for core region. Due to space restriction, we couldn't show data of genotyping methods for each paper in Table 1.
6.2. Prevalence of HCV Genotypes and Subtypes in Iran Provinces and Cities
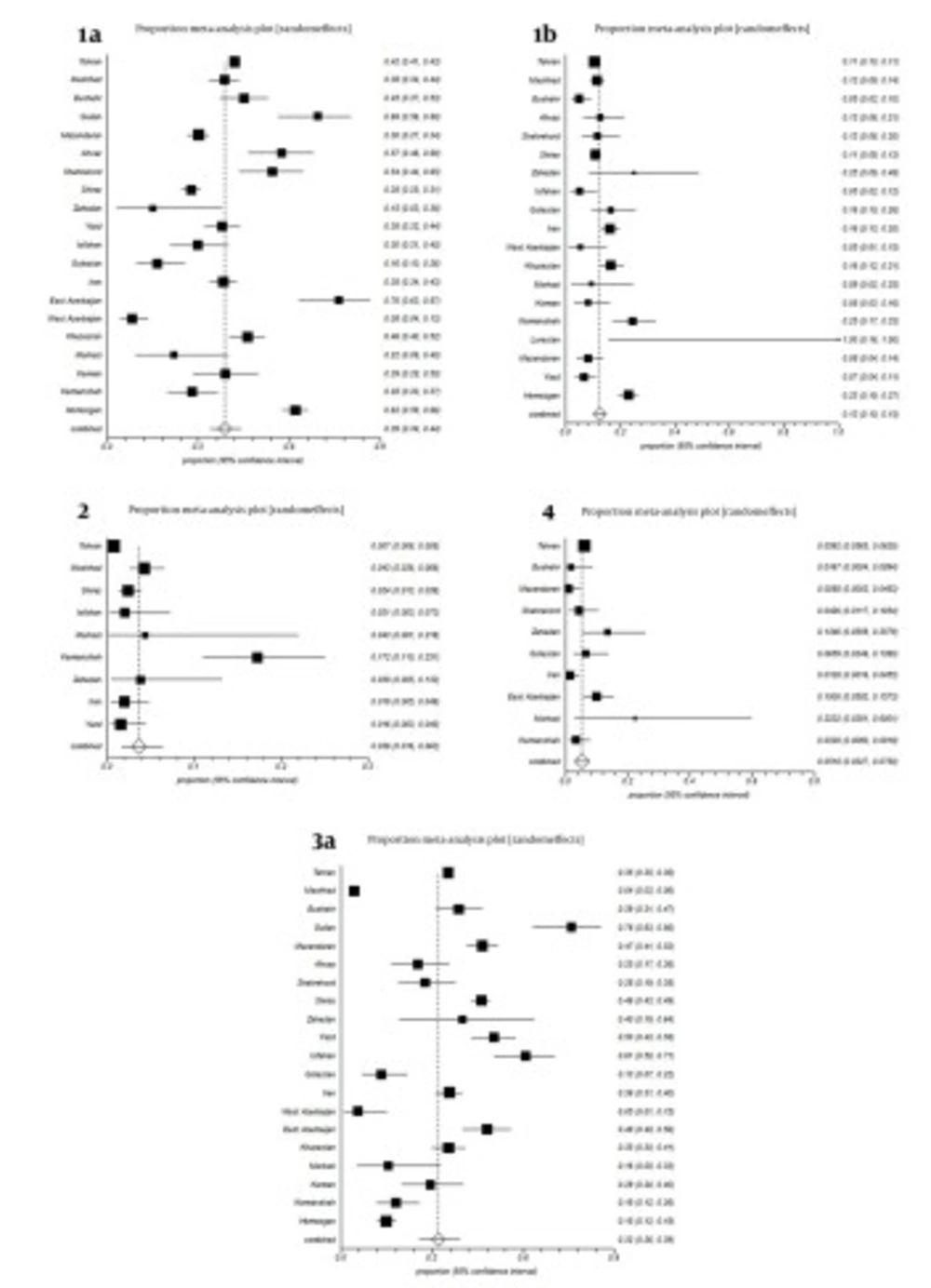
All studies were divided into 20 groups according to the place of sampling. Some studies were reporting HCV genotypes from all of Iran without separating data according cities or provinces; we included them in a separate group named, Iran (Table 1, Figure 2). Estimated pooled prevalence of HCV genotypes in patients distributed in provinces and cities of Iran were as follows; Subtype 1a was the predominant genotype with a rate of 39% (34-44%, 95% CI); followed by subtype 3a, 32% (26-39%, 95% CI); subtype 1b, 13% (10-15%, 95% CI); genotype 4, 5.18% (3.27-7.5%, 95% CI); and genotype 2, 3.6% (1.6-8.3%, 95% CI), respectively. Untypeable HCV was 0.3% (95% CI: 0.07-0.16%). Remnants were mixed genotypes 1.96%, genotype 5, 0.056% and other subtypes (Table 1, Figure 1). Genotype 6 was reported only in one study with prevalence rate 1 (0.7%) with subtype 6a (60).
Estimated overall prevalence of HCV genotypes was variable in cities and provinces of Iran. For example, subtype 1a was maximum in East Azarbyjan, 76% (95% CI: 63-87%) and Guilan, 69% (95% CI: 56-80%) respectively; but minimum in West Azarbayjan, 8% (95% CI: 4-13%). Prevalence of subtype 3a was variable in cities and provinces of Iran, for instance, maximum in Guilan, 76% (95% CI 63-86%) and Isfahan, 61% (95% CI, 50-71%) yet, minimum in West Azarbyjan, 5% (95% CI, 1-15%). Prevalence of subtype 1b was 13% (95% CI: 10-15%) in Iran, while in Kermanshah, Zahedan and Hormozgan, it was 25% (95% CI: 17-33%), 25% (95% CI: 9-49%), and 23% (95% CI: 19-27%) respectively. Prevalence of genotype 2 was 0.7% (95% CI: 0.06-0.8%) in Tehran, while in Kermanshah it was 17.2% (95% CI: 11-25.1%) (Figure 2).
6.3. Time
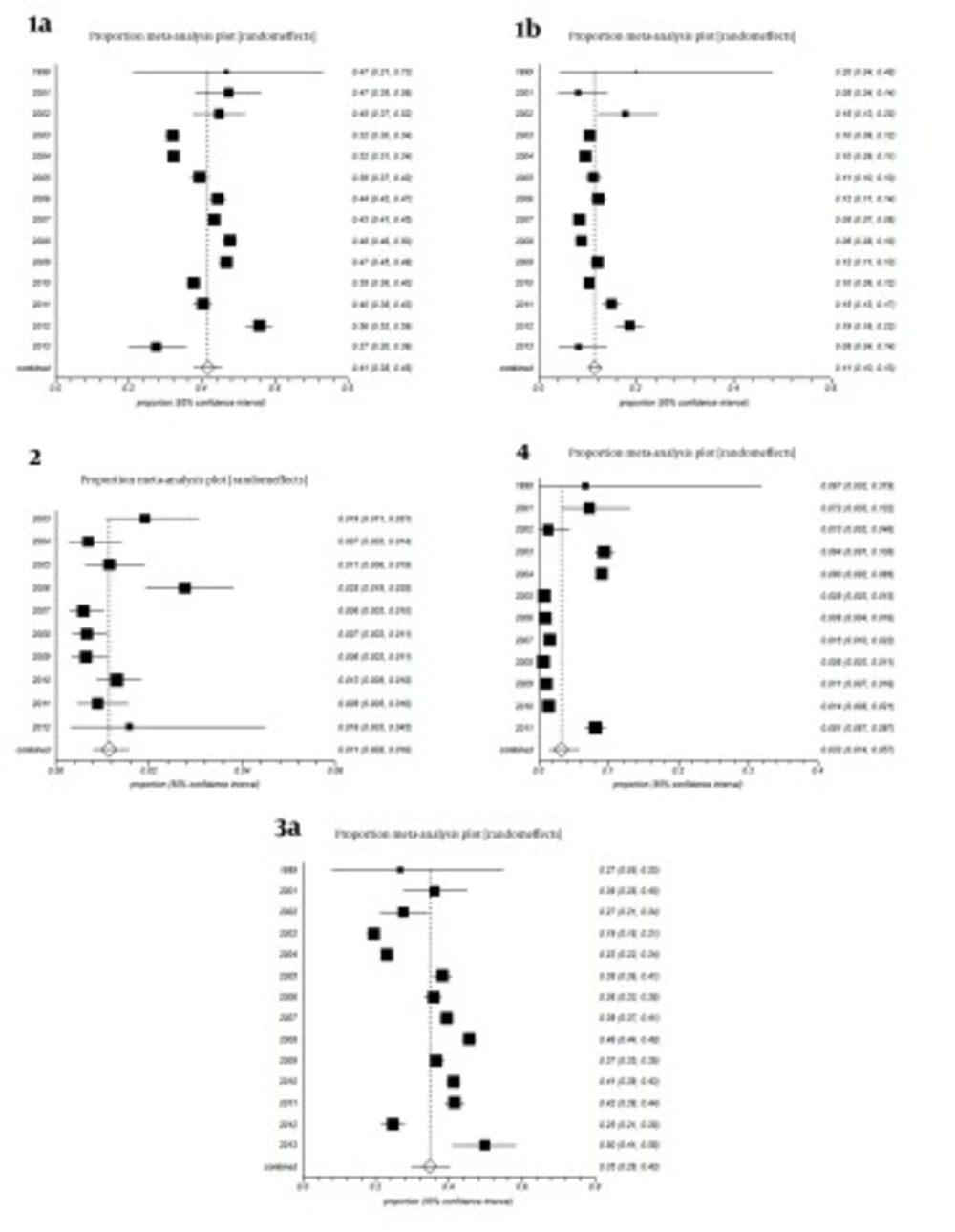
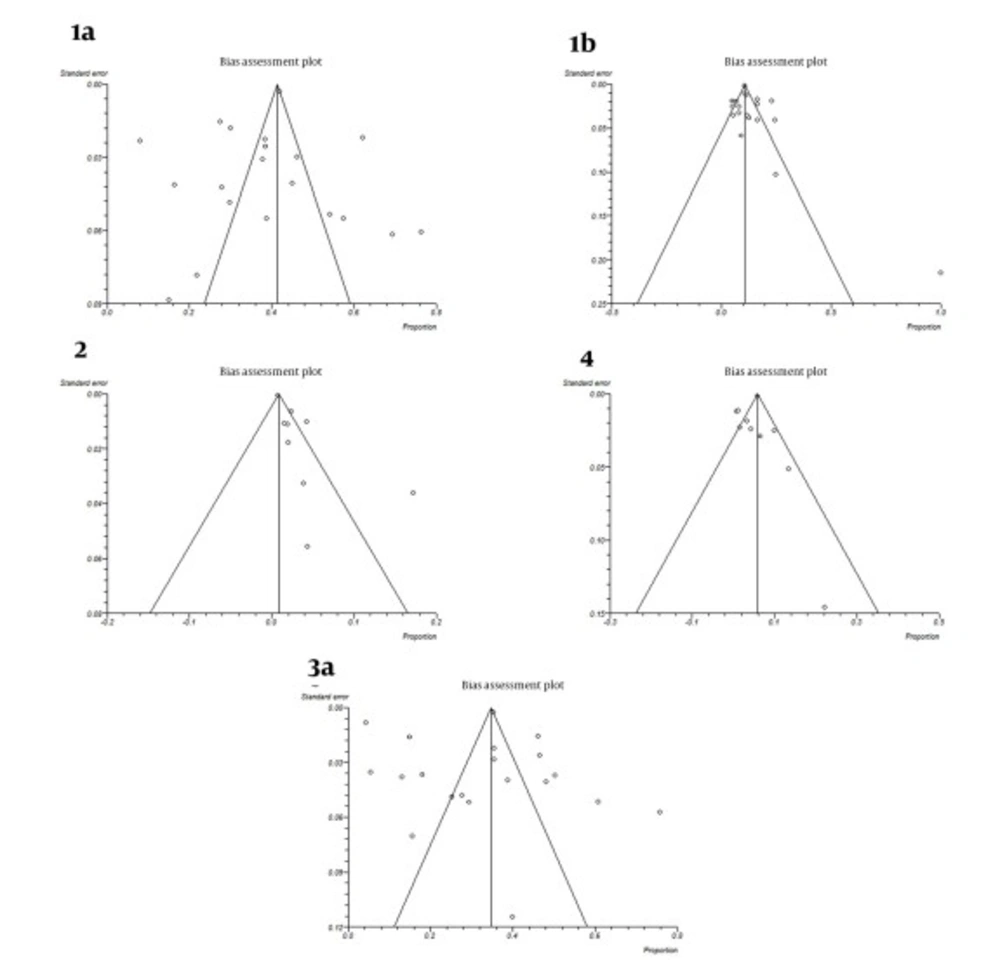
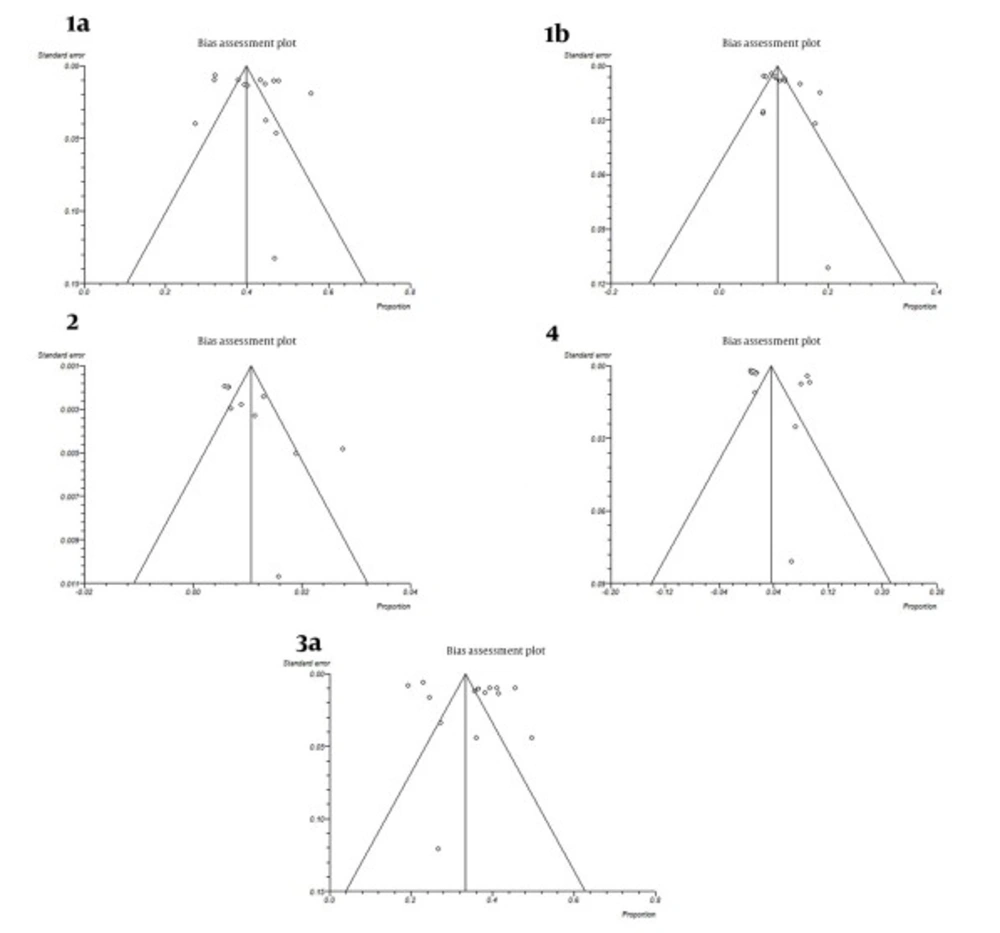
All studies were divided into 14 groups according to the year of study. The estimated annual prevalence rates of HCV genotypes over years are shown in Figure 3. Funnel plots for detecting biases in published papers are presented in Figures 4 and 5.
7. Conclusions
The frequency of HCV genotypes in different geographic regions is variable and changes over time, depending on the study population, the route of infection and virus mutation (3, 11). Considering countries around Iran, the predominant HCV subtype is 1b in Turkey, Russia, Moldova and Uzbekistan (10), 4 in Saudi Arabia, Iraq, Qatar, Bahrain, Kuwait and Yemen (12), and 6a in China (3). The predominant subtype is 3a in Pakistan (6) and India (33). Despite the high number of visitors from and to Arab countries (Iraq, Qatar, Bahrain, Saudi Arabia, Kuwait) with predominant genotype 4, and visitors from north neighboring countries such as Tajikistan and Turkmenistan with the predominant subtype 1b; subtype 1a is dominant in Iran. It is probable that differences in the population, routes of transmission, and socioeconomic factors may explain this variation.
The prevalence of genotype 4 in Iran is lower compared to the majority of Middle Eastern countries including the neighboring Arabic countries. In Iran genotype 4 is prevalent in patients on hemodialysis and this is related to communication with neighboring countries such as Iraq and Saudi Arabia during the pilgrims and Hajj ceremony. There are no reliable reports about HCV genotypes in Afghanistan, the eastern neighbor of Iran. During the past few years, due to the political situation of Afghanistan, a considerable number of immigrants came to Iran. These immigrants usually do many trips between Iran, Afghanistan and Pakistan (11).
Only one study has reported HCV genotype 5 (3.4%) in their study population (25), which can be a result of genotyping mistake and should be confirmed by DNA sequencing. Genotype 6 was reported only in one study with a prevalence rate of 1/ 142 (0.7%) with subtype 6a in one patient who had referred to the Gastroenterology Department of Taleghani Hospital (Tehran, Iran) between June 2007 and June 2012 (60). It seems that this patient was infected by HCV in a country other than Iran.
In one study that was performed recently on 11561 chronic HCV infected patients in Iran, the results revealed that, 1a was common (44.9%) followed by 3a (39.6%) and 1b (11.3%) (12). Distribution of HCV genotypes in our population is similar to the pattern on HCV in northern Europe, where genotypes 1, 2 and 3 are more frequent (25, 33).
In the last decade, an increase in HCV genotype distribution has been reported in many countries, for example there has been an increase in the prevalence of the 3a, 1a, and 4 (12). Documents have indicated that HCV infection has risen dramatically in the recent years in the Middle East region. This might be related to changes in the main route of transmission (61). Subtype 1a and 1b are the dominant genotypes in older and 3a in young patients in Iran (25). The high prevalence of subtype 3a in young patients suggests an increased number of IVDU as the main route of HCV transmission. In addition, recent studies have indicated that there is an increase in the frequency of 3a in the younger population of Iran, Germany, Serbia, Montenegro, and Slovenia; this is because the main route for transmission of HCV is IVDU, due to sharing of unsterilized needles and syringes. Therefore, young male individuals are more susceptible to acquire HCV infection (12).
Considering forest plots in Figure 3, our results showed that trend of hepatitis C virus genotypes 1a and 1b was fluctuating from 1999 to 2013, with a sinus shape, reaching maximum levels in 2012 and minimum levels in 2013; the reasons of these fluctuations are unknown. The overall trend of genotypes 2 and 4 was decreasing yet there was an increase for genotype 2 in 2012 and genotype 4 in 2011. The overall trend of subtype 3a decreased from 2001 to 2003 yet increased from 2004 to 2011, while it suddenly decreased in 2012 followed by a sharp increase to maximum levels in 2013. The reasons of these fluctuations is unknown. It must be noted that changes in the route of transmission and easy travelling in the recent years are the probable cause. Further researches are needed to find definitive answers.
Mixed HCV infection with more than one genotype is possible. Numerous reports suggest that HCV infection with one genotype does not prevent becoming infected with other HCV genotypes. Multiple exposures to HCV may occur from time to time. Mixed infections may lead to severe disease, poor response to antiviral therapy or relapse after treatment. Studies have reported that mixed infection with two genotypes of HCV occurs in approximately 1% of HCV infected patients, and 1.6 to 31% of patients with multi-transfused hemophilia. In one study multiple HCV subtypes (1a and 3a) were detected in 2.5% of the patients (12). In the present study the prevalence of mixed HCV infection was 1.96%, which is compatible with previous reports.
A funnel plot is helpful for detecting biases around the line of identity in the meta-analysis. In our study the bias assessment plot was symmetric for subtypes 1a, 1b, 3a and genotype 4, yet asymmetric for genotype 2 in papers reporting HCV genotypes in Iranian cities (Figure 4). Research insufficiency may be the reason of bias for genotype 2. Also, according to study periods, bias assessment plots were symmetric for genotypes or subtypes 1a, 1b, 2, 4 and 3a (Figure 5).
Forest plots of the present study indicated heterogeneity in the published papers (Figures 2 and 3). On the other hand, forest plots showed that, as the sample size increases confidence intervals of estimated prevalence become narrower. Thus, large sample sizes increase the precision of the study. In addition, samples collected from some parts of Iran were not adequate for obtaining firm conclusions.
A proportion of HCV in Iranian patients was reported untypeable. Presence of untypeable HCV samples in some studies indicates the inability of genotyping methods to discriminate HCV genotypes. To be able to identify untypeable HCV genotypes, sequencing and phylogenetic analysis of viral genome is needed for characterization. Thus, the future goal of researches should be to sequence genomes of untypeable samples. However, in some papers of the present study genotypes of HCV were detected by sequencing.
The transmission of HCV occurs mainly through infected blood, blood products, and contaminated devices, such as syringes and needles. Today, intravenous drug use is an important route of HCV transmission (4, 61). Precise data about the pattern of HCV genotypes and continuous monitoring of the genetic diversity of virus isolates, especially among high-risk individuals (hemophilia, thalassemia, hemodialysis, multiply transfused patients and intravenous drug use) is essential for understanding HCV epidemiology (33). The evaluation of HCV risk factors among patient populations is important. Most studies did not report on HCV genotypes in patient populations completely. Because of this defect in data, we were unable analyze the prevalence of HCV genotypes according to patient’s population.
Male gender was reported as an independent predictive factor for HCV infection in previous studies (61). The findings of a recent study showed that there was no association between having a variety of HCV genotypes and gender, level of education, risk factors, job, income, HIV infection, HBV infection, IV drug abuse and presence of underlying diseases (3). In our study most of the HCV infected patients were male (Table 1). However, most studies have not reported prevalence of HCV genotypes according to gender and above-mentioned factors separately. Thus, we were unable analyze the prevalence of HCV genotypes according to gender and risk factors.
A range of studies on the prevalence of HCV genotypes in Iranian patients have been conducted. A meta-analysis of these prevalence data would be important for public health policy making and planning of clinical services addressing the needs of these infected people. The advantage of this meta-analysis is that it merges a large sample size and long study period, which help draw a reliable estimate of HCV genotypes distribution in Iran. Our study had a number of limitations that should be mentioned. First, different HCV genotyping methods were used by various studies and this may impact the obtained results of this meta-analysis. Second, although study populations were all Iranian patients, yet some populations were special patient groups and including them in a meta-analysis is somewhat challenging. Furthermore, data limitations of some studies did not allow their analysis.
In conclusion, our study showed that the most frequent subtypes of HCV in Iran were 1a, 3a and 1b, respectively. This frequency differs in cities and provinces of Iran and neighboring countries. Also, this meta-analysis reported on the overall estimate for distribution of HCV genotypes in Iran. Therefore, due to the possibility of changing dominant viral genotypes in communities based on migrations and viral genome mutations, it is important to determine distribution of HCV genotypes in different geographical areas and its trend with time for epidemiological and patient management purposes.