1. Background
Nonalcoholic steatohepatitis (NASH) has been shown to be associated with obesity, diabetes and hyperlipidemia (1). The disease pathophysiology is not fully clear and seems to be multifactorial. Current models propose the “two hit hypothesis”. First, lipids accumulate in hepatocytes and then trigger inflammation by a variety of mechanisms (2). Insulin resistance, lipid peroxidation, oxidative stress, endotoxins and cytokines are possible mechanisms in the pathogenesis of NASH. In addition, some genetic backgrounds may have considerable effects on the clinical course and prognosis of disease (3).
Interleukin 8 (IL8), a family member of the chemokine, has a role in induction and amplification of inflammatory processes (4). Increased plasma IL 8 levels were detected in NASH patients (5-7). Therefore, it may have a crucial role in NASH disease in which inflammation is a substantial pathophysiological feature. The (-251 A/T) polymorphism in IL 8 gene’s promoter region is the only one influencing gene expression (8).
Interleukin 6 (IL 6), as a cytokine, has pleiotropic features, takes part in regulation, proliferation, differentiation and various activities of cell types. It also has an important part in neuro-endocrine and homeostasis of immune system (9). Dysregulated IL 6 production is implicated in the pathology of several disease processes such as multiple myeloma, arthritis, diabetes, atherosclerosis and hepatocellular carcinoma (HCC) in patients with chronic hepatitis B (10-13). In 5' upstream of IL 6 (-174 C/G locus) a localization of C/G polymorphism has been shown (14). This basic modulation has been correlated to various IL 6 plasma values and proportion of IL 6 gene transcription in healthy subjects (10). Furthermore, increased plasma IL 6 levels were found in patients with NASH in a pilot study (6). Increasing scientific facts indicate that interracial differences and genetic factors have an impact on the natural history of chronic liver disorders. It has been concluded that gene polymorphisms affect the advancement of liver fibrosis among patients infected with hepatitis C virus (HCV), autoimmune chronic cholestasis and autoimmune liver diseases (15). The roles of genetic factors in NASH have been estimated in several studies (10, 11, 16). Several candidate genes relevant to the “second hit hypothesis” have been preliminarily examined. A polymorphism in manganese superoxide dismutase (MnSOD), a gene limiting mitochondrial oxidative stress, was reported to be enriched as a cohort in patients with NASH (17). Polymorphisms in two genes, angiotensinogen and transforming growth factor-β1, have been associated with greater fibrosis in morbidly obese subjects undergoing bariatric surgery (18). A Japanese study revealed that polymorphisms of Interleukin 1-beta and beta-3 adrenergic receptors affect the development of NASH (19).
2. Objectives
We aimed to investigate whether IL 6 and IL 8 gene polymorphisms play role in identification and pathophysiology of NASH and to evaluate the roles of the promoter (-174 G/C) of IL 6 and promoter (-251 A/T) of IL 8 polymorphisms.
3. Patients and Methods
3.1. Patients
Consecutive participants with biopsy proven NASH who had persistently elevated liver enzymes and hepatosteatosis on ultrasonography, in the absence of any causes of elevated aminotransferase levels, such as viral hepatitis, sclerosing cholangitis, autoimmune hepatitis, primary biliary cirrhosis, Wilson’s disease, hemochromatosis, alpha- 1-antitrypsin deficiency, malignancy and drug-induced liver disease were enrolled in the study. Excessive alcohol consumption of (> 20 g/day) was also considered as exclusion criteria. The healthy control group included consecutive age- and gender-matched individuals who had normal aminotransferase levels and normal abdominal ultrasonography and without any illness, alcohol consumption, drug or herbal substance usage, history of previous liver diseases and with negative results for viral hepatitis markers.
Demographic and clinical data of patients were collected and registered in a database by the same clinician to prevent bias. Anthropometric measurements, including height, body weight and body mass index (BMI), were recorded. BMI was calculated as weight (in kilograms) divided by the square of height (in meters) as kg/m2 in all patients and controls.
3.2. Evaluation of IL Genotyping
Blood samples were taken in bottles containing 72 μL 7.5% concentrations of K3-EDTA (BD, USA). For DNA isolation, 200 μL of blood from the samples was used and purificated by EZ-DNA Genomic DNA Isolation Kit (Biological Industries, Israel). Polymerase chain reactions (PCRs) were performed with appropriate primers after genomic DNA was obtained from the peripheral blood of patients and control subjects. For determination of IL 6 gene -174G/C (rs1800795), and IL8 -251 A/T (rs4073) polymorphisms, F-5’-TTCGTGCATGACTTCAGCTTTAC-3’ R-5-‘AGCCTCAGACATCTCCAGTCCT-3’ and F-5-‘CCATCATGATAGCATCTGTA-3’ R-5’-CCACAATTTGGTGAATTATTA-3’ primers were selected, respectively. Both PCR reactions consisted of 16mM (NH4) 2SO4, 67 Mm, TrisHCL pH 8.8, 0.01%Tween-20, 1.5 Mm MgCI2, 200 μM for each deoxynucleotide triphosphate, 0.026 U/ μL TaqDNA polymerase enzyme (Bioron, Germany) and nearly 0.5 μg mold of DNA was put in each tube. PCR conditions regarding both of the polymorphisms were performed as follows: denaturation at 94°C for five minutes, followed by 30 cycles of denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds and extension at 72°C for 30 seconds, with a final extension at 72°C for five minutes. We used a control without DNA to check for contamination. Two percent agarose gel revealed a PCR product of 330-bp for the -174G/C polymorphism and a 174-bp for IL 8 -251 A/T polymorphism.
To genotype -174G/C polymorphism, a 330 bp PCR product was digested with 10 units of SfaNI restriction enzyme at 37°C for overnight. The results were evaluated with two blind observers after electrophoresed in a 3% agarose gel. The presence of restriction sites on both alleles was digested in fragments of 182 and 148 bp were labeled as GG genotype; whereas, the absence of restriction sites, gave one 330 bp fragment revealed as CC genotype.
Regarding the genotype pf -251 A/T polymorphism of IL 8 gene, a 174 bp PCR product was digested with 10 units of AseI restriction enzyme at 37°C for overnight. The results were evaluated with two blind observers after electrophoresed in a 3% agarose gel. The presence of restriction sites on both alleles was digested in fragments of 152 and 22 bp were labeled as AA genotype; whereas, the absence of restriction sites, gave one 174 bp fragment revealed as TT genotype.
3.3. Liver Histopathological Evaluation
Percutaneous liver biopsy was performed using a 16-G disposable needle by a skilled clinician. All liver biopsy specimens included 10-12 or more complete portal tracts and were at least 20-25 mm in length assessed by an experienced hepatopathologist blinded to clinics and anthropometric data of patients. Tissue specimens were fixed in formalin, embedded in paraffin and prepared with Hematoxylin/Eosin and Masson’s trichrome stains to assess the morphology and fibrosis of the liver.
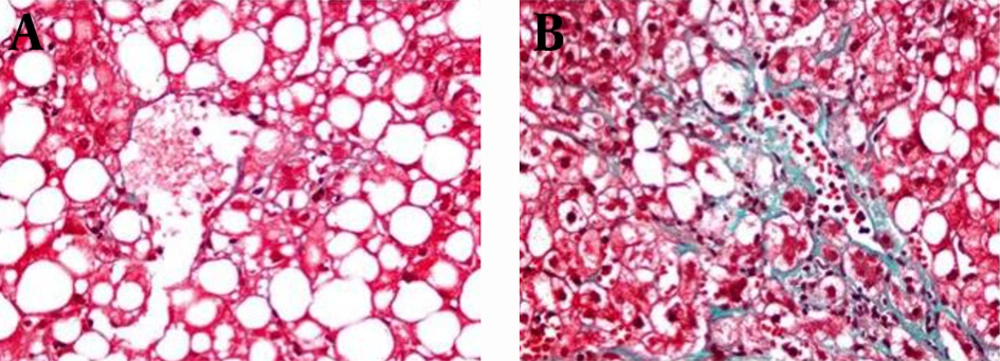
Diagnosis of NASH was made according to the Brunt’s Criteria (20). Histological features were graded in accordance with the NAFLD scoring system (NAS) and the National Institute of Diabetes and Digestive and Kidney Diseases NASH Clinical Research Network (21). Hepatic steatosis was according to steatosis percentage; 5-33%, 33-66% and > 66% denoting scores 1, 2 and 3, respectively. Lobular inflammation was identified as the entire assessment of all inflammations; no foci as score 0, < 2 foci per x200 field as score 1, two-four foci per x200 field as score 2, > 4 foci per x200 field as score 3. Ballooning scoring was termed as; score 0 no ballooning; score 1 as few and score 2 existence of numerous ballooning of hepatocytes. Liver fibrosis was staged as follows; stage 0, no fibrosis; stage 1, perisinusoidal or periportal fibrosis; stage 2, perisinusoidal and portal/periportal fibrosis; stage 3, bridging fibrosis and stage 4 as cirrhosis. Histopathologically, total NAS was calculated as a sum of steatosis (1–3), lobular inflammation (0–3) and ballooning (0–2). NAS ≥ 5 was identified as definite NASH and all patients were diagnosed as definite NASH. Patients were classified as mild and significant fibrosis groups. While, mild fibrosis group was characterized as fibrosis score < 2 pathologically represented in Figure 1 A and significant fibrosis group was characterized as fibrosis score ≥ 2 shown in Figure 1 B (22).
3.4. Ethics
This study was performed according to the ethical guidelines of the 1975 Declaration of Helsinki, as updated in 2008. The local ethics committee of Gazi University School of Medicine approved the research protocol by the number of 244.2006.16.6. Informed and signed consents were obtained from all patients and healthy controls.
3.5. Statistical Analysis
Statistical analysis was performed using SPSS version 21 (Statistical Package for Social Science; Chicago, IL, USA). The variables were investigated using visual (histograms, probability plots) and analytical methods (Kolmogorov-Smirnov/Shapiro-Wilk’s test) to determine whether they were normally distributed. Descriptive analysis was presented as frequencies for ordinal variables and as mean ± standard deviation for normally distributed continuous variables. The comparisons of non-normally distributed continuous variables were performed using Mann-Whitney U test. Chi-square test or Fisher’s exact test, where appropriate, was used to compare the groups. The comparison of normally distributed continuous variables was performed using Student t-test. The comparisons of multiple groups were made by Kruskal-Wallis test and Bonferroni correction was performed where appropriate. P < 0.05 was noted statistically significant for all analyses.
4. Results
Thirty-eight consecutive patients with biopsy proven NASH and 38 age- and gender-matched voluntary healthy controls were recruited in the study compatible with inclusion and exclusion criteria. Age and gender of groups were comparable with each other. NASH patients had higher BMI than healthy controls 929.3 ± 2.3 vs. 27.8 ± 2.1, P = 0.007). The main baseline characteristics of patients and healthy controls are shown in Table 1.
The comparisons of frequencies of the two genotypes G/C and G/G of IL 6 between NASH group and healthy controls were 39.5%, 60.5% vs. 53.6%, 46.4%, respectively, but there was no statistically significant differences between the two groups (P > 0.05). The frequencies of genotypes of IL 8 among NASH group were 47.2%, 44.6% and 8.2% for T/T, A/T, and A/A, while among healthy controls were 50%, 28.6% and 21.4%, respectively, (P > 0.05). The frequencies of distribution of IL 6 and IL 8 gene polymorphisms between patients with NASH and healthy controls are shown in Table 2. Liver biopsy samples were consistent with histopathological diagnosis of NASH and NAS scores were ≥ 5 in all patients. According to fibrosis scores there were 27/38 (71%) patients in mild fibrosis group, while there were 11/38 (29%) in significant fibrosis group. From all patients, 4 (10.5%) were in F0, 23 (60.5%) in F1, 6 (15.8%) in F2, 4 (10.5%) in F3 and 1 (2.7%) in F4.
To further investigate the role of genetic polymorphisms and genotypes between fibrosis groups we compared the frequencies of -174 C/G polymorphism in the IL 6 gene, -251 A/T polymorphism of IL 8, G/G and G/C genotypes of IL6 between the groups, but there was no statistically significant difference (P > 0.05). However, the A/A genotype was significantly higher in patients of significant fibrosis group than the mild fibrosis group (27.3% vs. 0%), respectively and (P = 0.0016). Comparing T/T, T/A and A/A genotypes of IL 8 gene between fibrosis groups is shown in Table 3.
| NASH Patients (n = 38) | Healthy Controls (n = 38) | P Value | |
|---|---|---|---|
| Age, y | 45.5 ± 10.2 | 46.5 ± 8.5 | NS |
| Gender, Female/Male | 14/24 | 14/24 | NS |
| BMI, Kg/m² | 29.3 ± 2.3 | 27.8 ± 2.1 | 0.007 |
a Abbreviations: BMI, body mass index; NASH, nonalcoholic steatohepatitis; NS, not significant.
b Data are expressed as mean ± SD.
| Genotypes | NASH Patients | Healthy Controls | P Value |
|---|---|---|---|
| IL 6 | |||
| G/G | 23 (60.5) | 18 (46.4) | NS |
| G/C | 15 (39.5) | 20 (53.6) | NS |
| IL 8 | |||
| T/T | 18 (47.2) | 19 (50) | NS |
| T/A | 17 (44.6) | 11 (28.6) | NS |
| A/A | 3 (8.2) | 8 (21.4) | NS |
a Abbreviations: NASH; non-alcoholic steatohepatitis, NS; not significant.
b Data are presented as No. (%).
a Abbreviations: NASH; non-alcoholic steatohepatitis, NS; not significant.
b Data are presented as No. (%).
c Mild fibrosis: Fibrosis score < 2.
d Significant fibrosis: Fibrosis score ≥ 2.
5. Discussion
In this research, we investigated IL6 and IL8 gene polymorphisms in NASH patients and healthy controls and attempted to define their roles in the pathogenesis and severity of disease. Our results indicated that polymorphisms in IL 6 and IL 8 genes had no effect on the identification of NASH as there was no difference compared to healthy controls. However, the presence of A/A genotype of the IL 8 gene is associated with disease progression. Besides, polymorphisms of -174 C/G polymorphism in the IL 6 gen and -251 A/T polymorphism of IL 8 had no roles in the progression of liver fibrosis. The poorly understood pathogenesis of NASH seems to be multifactorial (2). In this study, we confirmed higher BMIs NASH patients. In a Japanese study, it was concluded that obesity is a substantial variable in the progression of NASH (19).
While environmental risk factors clearly affect the emergence and development of NASH, the diversity of phenotypes in persons with similar metabolic risk factors strongly implies a genetic contribution (23). A useful and commonly held theory to explain the pathogenesis of NASH is the “two hit” hypothesis (2). The “first hit” is characterized by initiation of hepatic steatosis and the “second hit” is characterized by increased intracellular oxidative stress that can be induced by numerous mechanisms, including endotoxin exposure, pro-inflammatory cytokines (23). Proinflammatory cytokines, IL6 and IL8, have been thought to be involved in the pathogenesis of NASH, but the underlying mechanisms have not been determined exactly yet (5-7). In this study, we investigated IL6 and IL8 gene polymorphisms in NASH patients and attempted to define their role in the pathogenesis and severity of disease. In our study, we assessed the role of IL 8 polymorphism in the “second hit” pathogenesis of NASH, but no statistically remarkable difference was seen between NASH patients and healthy individuals. Nevertheless, the A/A genotype was higher among patients with severe NAS. These observations support the idea that the A/A allele is profoundly involved in the NASH progression. IL8, a substantial cytokine, takes part in the inflammatory process. Bahcecioglu et al. found that serum IL8 values of NASH patients were higher than controls (5). Moreover, in another study IL8 was significantly higher in NASH patients than steatosis group (7). A study from China concluded that IL 8 may take part in the pathogenesis of NAFLD in Chinese patients and concluded that patients who had high ALT levels had higher IL 8 levels. Nevertheless, there was no difference among NASH patients (24). The -251A/T polymorphism in the promoter zone of the IL 8 gene affects its expression. The mutant allele “A” of the -251A/T polymorphism is less common, but its presence increases the production of IL 8 cytokine (8). A pilot study found that monocytes from NASH patients overproduced tumor necrosis factor (TNF), IL 6 and IL 8 and after lifestyle modification and vitamin E therapy, the levels of IL 8 remained elevated. This data indicates that a metabolic defect in these monocytes, which permits the overproduction of cytokines, may be responsible for disease pathogenesis. The metabolic defect can result from genetic defects.
IL6 is a multifunctional cytokine with a critical role in host defense, and has important features like stimulating the hepatic acute phase response to infection and injury. IL6 can be rapidly cleared from the plasma, so circulating levels of IL6 are largely regulated by its expression (10). A C/G polymorphism of IL 6 has been related to different IL 6 serum plasma values and transcription rates of IL 6 gene among healthy individuals (10, 14). Furthermore, plasma IL 6 values were increased in NASH patients in a pilot study (6). In our study, the role of IL 6 polymorphism in the “second hit” pathophysiology of NASH was examined, but no statistically remarkable difference was found between NASH patients and healthy controls. Some studies concluded that IL6 polymorphisms affect histopathological progression of HCV, hepatic insulin resistance, inflammation and occurrence and development of HCC (12, 13, 25-27).
In our study, among NASH patients, there was not a difference between the fibrosis groups regarding IL6 and IL8 gene polymorphisms. NASH may progress to liver fibrosis, liver failure and eventually death. However, not all individuals exposed to a similar causal agent develop the same degree of liver fibrosis. Besides, there is very little information about factors affecting fibrosis development in NASH (28). While some studies showed that HFE C282Y heterozygosity and PNPLA3 gene are associated with faster fibrosis advancement, the others have not concluded these outcomes (15, 29). Besides, the impact of interracial differences on the pathophysiology of this common disease should be investigated. In conclusion, we showed that IL6 and IL8 gene polymorphisms had no effect on NASH pathogenesis and progression of liver fibrosis; however, the presence of A/A genotype in IL 8 gene is associated with disease progression. Genetic factors influencing the progression of liver fibrosis among NASH patients need further investigation in different racial populations.