Fulltext
Hepatitis C virus (HCV) infection is a major cause of chronic hepatitis, liver cirrhosis and hepatocellular carcinoma (1). The interaction between HCV and host innate immunity plays a key role in the immunopathogenesis of HCV disease. Innate immunity is the first line of defense against viral infections. The host innate immune system recognizes pathogens and responds to their stimuli mainly through pattern recognition receptors (PPRs). Toll-like receptors (TLRs) are key sensors that recognize viral pathogen-associated molecular patterns (PAMPs) during viral infection (2). Several TLR members play a critical role in recognition of viral nucleic acids (3). Among these TLRs, TLR3 has a crucial role in virus-mediated innate immune responses (4, 5), as it recognizes dsRNA (6), which either constitutes the genome of one class of viruses or is generated during the life cycle of many viruses, including HCV (4, 5, 7). Sensing through TLR3 activates IFN signaling pathway and induces the production of type І IFNs (IFN-α/β). IFN- α/β have been recognized as the first line of TLR3 activation-mediated antiviral response (8). Therefore, activation of TLR3 signaling pathway in viral target cells could inhibit virus infections such as herpes simplex virus-1 (HSV-1) (9), HIV (10) and HCV (7).
Although cellular and humoral immune responses are present during acute and chronic HCV infection, they are ineffective in eradicating the virus (11). Most HCV-infected subjects develop chronic infection, suggesting that HCV has evolved the strategies to overcome or evade host immune responses (12). Studies (12) of HCV-host interactions revealed that HCV can use several approaches to compromise the host immune response during viral infection. HCV NS3/4A protease is able to impair TLR3 signaling by cleaving the Toll-IL-1 receptor domain-containing adaptor inducing IFN-β (TRIF) adaptor protein and block RIG-I signaling by cleaving the mitochondrial antiviral signaling protein (MAVS) of the mitochondria to inhibit IRF-3 activation and IFN-β expression (13-15).
Macrophages are phagocytic mononuclear cells of the innate immune response, which also participate in adaptive responses. Resident liver macrophages, Kupffer cells (KCs) constitute 15-20% of the total nonparenchymal cells in the liver (16). KCs sense pathogens and help to maintain the tolerogenic environment of the liver (16). Total macrophage numbers increase significantly in chronic HCV-infected liver, which represents a local proliferation of KCs and infiltration of bone-marrow-derived monocytes from peripheral blood (17). Macrophages are key regulators of early immune responses during viral infection of the liver. Macrophages determine cellular and cytokine milieu in HCV-infected liver and contribute to liver inflammation (18). Furthermore, macrophages influence HCV-induced liver fibrosis, cirrhosis and hepatocellular carcinoma (18). However, the role of macrophages in liver innate immunity against HCV replication is poorly understood.
In the present study, we aimed to investigate the role of macrophages in liver innate immunity against HCV replication.
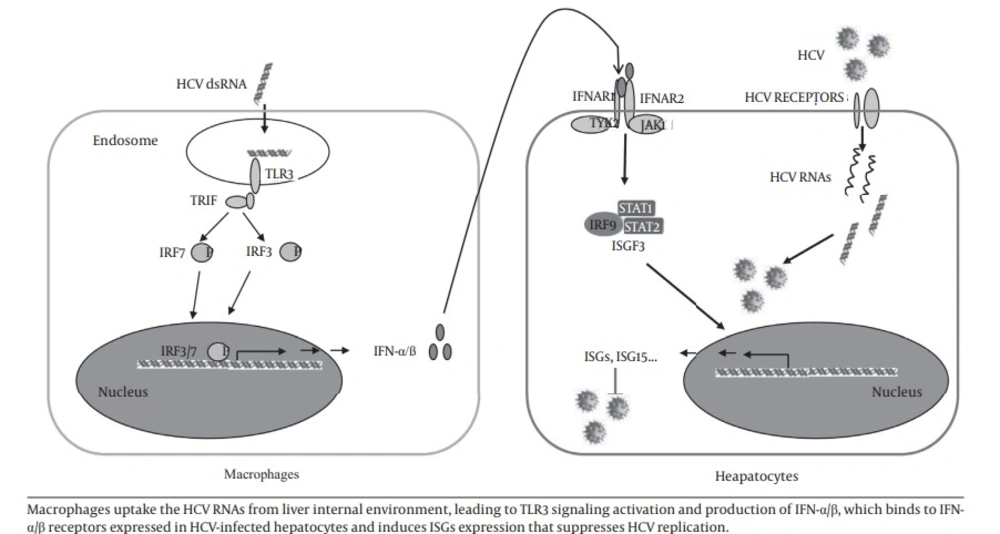
In this study, we investigated the role of macrophages in control of HCV replication in hepatocytes. Four HCV dsRNAs (Core, E1-P7, NS-3’NTR and NS5A) were used to investigate the effect of HCV genome on the activation of MDM. Studies (24-26) showed that HCV RNA is highly structured and contains ds regions in various portions of the genome, such as the 5’- and 3’-NTRs and the core- and NS5B-coding regions. Other studies also showed that these four HCV dsRNAs could induce chemokine and inflammatory cytokine expression in hepatocytes (25) and IFNs expression in human type 2 myeloid dendritic cells (26). We showed that HCV dsRNA-activated MDM had the ability to suppress HCV replication in hepatocytes. We observed that HCV dsRNA induced IFN-α/β expression in macrophages. IFN-α/β is the first line of innate immunity against viral infections. We further demonstrated that IFN-α/β were involved in this macrophages-mediated HCV inhibition, which was evidenced by the observation that antibody to type I IFN receptor (IFNAR2) could neutralize the macrophages-mediated anti-HCV effect. In addition, the role of IFN-α/β in macrophages-mediated anti-HCV activity is supported by the observation that macrophages SN treatment induced the expression of ISG15, ISG56, OAS-1, OAS-2, MxA and Viperin in HCV-infected Huh7 cells. These results indicate that macrophages may be a key regulatory bystander, participating in host innate immunity against HCV replication using a type I IFN-dependent mechanism.
Furthermore, HCV dsRNA induced the expression of TLR3 and IRF-7 in macrophages, the key regulators of the IFN signaling pathway. As an important PPR, TLR3 recognizes dsRNA that either constitutes the genome of one class of viruses or is generated during the life cycle of viruses, including HCV dsRNA (20, 27). During the early stages of HCV infection, KCs and infiltrated macrophages are exposed to free viral nucleic acids and proteins. TLR3 expressed in macrophages likely recognizes HCV via phagocytosis and becomes activated, then induces the expression of IFN-α/β and contributes to viral inhibition (18). Here, we used synthesized HCV dsRNA to activated macrophages. We showed for the first time that HCV dsRNA can induce IFN-α/β expression in macrophages in vitro. It is most likely that HCV dsRNA induces IFN-α/β expression by activating TLR3 signaling pathway in macrophages.
Because of a lack of proofreading by the RNA-dependent RNA polymerase (RdRp) and high replication level, HCV presents a high degree of genetic variability. According to this genetic variability, classification of genotypes, subtypes, isolates and quasispecies is allowed (28). HCV variability and of quasispecies dynamics play important roles in infection transmission, mechanisms of chronicity and resistance to antiviral therapy (28). Thus, HCV genetic variabilities may potentially affect the activation of TLR3 signaling pathway in macrophages. One weakness of the current study was that we only used HCV dsRNAs generated from single HCV isolate. Future studies are needed to investigate the effect of HCV dsRNA of different genotypes on the TLR3 activation in macrophages.
As important immune cells, macrophages are key regulators of the early immune responses during viral infection in the liver. Macrophages play very important roles in the immunopathogenesis of HCV-related diseases. It was reported that macrophages participate in determining cellular and cytokine milieu in HCV-infected liver, contribute to liver inflammation and influence HCV-induced liver fibrosis, cirrhosis and hepatocellular carcinoma (18). Recent studies (29) showed that HCV can infect primary human macrophages in vitro, inducing tumor necrosis factor-α (TNF-α) and interleukin 8 (IL-8) expression. Other studies (30) showed that HCV can infect monocytes, macrophages and dendritic cells (DCs) in vivo. It was also showed that high concentrations of HCV particles could stimulate macrophages to express TNF-α and promote HCV entry into polarized hepatoma cells (31). Hepatic macrophages (KCs) link HCV infection with liver inflammation and disease through producing IL-1β (32). It is also showed that serum IL-1β levels are elevated in patients with chronic hepatitis C compared to healthy controls and KCs are the primary cellular source of IL-1β (32). IL-1β drives proinflammatory cytokine, chemokine and immune-regulatory gene expression networks with HCV disease severity (32). KCs can engulf apoptotic bodies of hepatocytes and stimulate death ligand and cytokine expression, which promote liver inflammation and fibrogenesis (33). Furthermore, HCV core and NS3 proteins can activate TLR2 signaling of macrophages and stimulate TNF-α and IL-10 production (34), and TLR1 and 6 are involved in this TLR2-mediated macrophages activation by HCV proteins (35). HCV also can activate inflammasome in monocytes and macrophages through TLR7 signaling pathway in an infection-independent manner (36). In our study, we demonstrated that HCV dsRNA-activated macrophages had the ability to suppress HCV replication in hepatocytes by producing IFN-α/β. Thus, macrophages play roles in both favorable and adverse responses to HCV infection in the liver.
Autophagy plays critical roles in many cellular processes including development, differentiation, survival and homoeostasis. It has been reported that autophagy plays an important role in the HCV-life cycle. HCV induces autophagy and uses autophagy-related proteins (Beclin 1, Atg4B, Atg5 and Atg12) for translation of viral mRNA to initiate its replication (37). Thus, the autophagy pathway can be a target for drugs development for HCV treatment. Although HCV is a hepatotropic virus, there are evidences to support the idea that HCV infection is a multifaceted disease, which causes many extrahepatic manifestations, such as mixed cryoglobulinemia, B-cell-derived non-Hodgkin’s lymphoma (NHL), diabetes mellitus, idiopathic pulmonary fibrosis, autoimmune thyroiditis, sicca syndrome, noncryoglobulinaemic nephropathies, glomerulonephritis and aortic atherosclerosis (38). Macrophage is a target cell of HIV. Because of similar transmission routes, HCV/HIV co-infection is a serious public health problem in high risk groups. It has been reported that antiretroviral therapy (ART) potentially increases the plasma HCV viral load among HCV/HIV co-infected individuals in an ethnic minority area in China (25). Taken together, chronic HCV infection is still a serious global health threat.
In conclusion, our study provided experimental evidences that macrophages may participate in liver innate immunity against HCV replication using a type I IFN-dependent mechanism (Figure 6).