1. Background
Hepatitis E virus (HEV), a member of the genus Hepevirus, is a non-enveloped virus with a positive-stranded RNA genome approximately 7.2 kb in length. HEV has been the cause of waterborne outbreaks of hepatitis in Asia and Africa and is a major cause of sporadic hepatitis in these regions. Acute HEV infection primarily affects young adults and is generally self-limiting and resolves in 1 - 6 weeks, except in women during late pregnancy, among whom 20% mortality has been reported; chronic HEV infection has recently been reported in transplant recipients (1). It has been hypothesized that zoonosis is involved in the transmission of HEV (2).
Hepatitis E virus and antibodies to HEV have been found in a wide variety of animals (3-6). Hepatitis E viruses were divided into 4 distinct genotypes: genotype 1, 2, 3, and 4. Genotypes 1 and 2 have been identified exclusively in humans, while genotypes 3 and 4 have been found in humans and several species of animals. Genotypes 1 and 2 have been isolated in Asia, Africa, and America; genotype 4 has been identified only in Asia; genotype 3 has been found almost worldwide (7, 8). Since 2000, genotype 4 HEV has become the dominant cause of hepatitis E disease in China (9, 10). Genotype 4 was proposed to be further classified into 24 subtypes (11). In the present study, we identified 52 genotype 4 hepatitis E viruses to subtype the HEV strains in eastern China. The relationship between HEVs from swine and human populations was also investigated.
2. Objectives
In the present study, we aimed to investigate the subtype of HEV prevalent in eastern China, and phylogenetically analyze the relationship between different HEV isolates prevalent in this area.
3. Materials and Methods
3.1. Samples
A total of 125 human serum samples, which were tested to be positive for anti-HEV IgM using commercial ELISAs (Wan Tai Pharmaceutical, China), were collected from local Centers for Disease Control and prevention (CDC) in Jiangsu Province by Jiangsu CDC from 2010 to 2013. A total of 290 swine fecal samples were obtained from pigs (aged 8 to 26 weeks old) in Zhenjiang City (80 samples) of Jiangsu Province, Anhui Province (90 samples) and Shanghai City (120 samples) from 2010 to 2012. Anhui Province and Shanghai City are two neighboring areas of Jiangsu Province. The three areas have the sample culture and interregional mobility of people is very common.
3.2. Detection of Hepatitis E Virus-RNA
All the fecal samples were converted to 10% (W/V) suspensions in 0.01 M Phosphate Buffered Saline (PBS) (pH 7.2 - 7.4). Fecal sample suspensions were then clarified by centrifugation at 10,000 g for 10 minutes and 100 μL aliquots of the clarified material was used for viral RNA extraction. Total RNA was extracted from 100 μL of serum sample or clarified fecal suspension, using Trizol (Invitrogen, USA). The viral RNA was finally dissolved in 20 μL RNase-free water. Reverse transcription was performed using the TaKaRa RNA polymerase chain reaction (PCR) kit (TaKaRa, Japan) according to the manufacturer’s protocol. Polymerase chain reaction was performed as described elsewhere (12). Briefly, a 348 bp segment of open reading frame 2 (ORF2), was amplified using primers E1 (5’-AATTATGCYCAGTAYCGRGTTG-3’) and E2 (5’-CCCTTRTCYTGCTGMGCATTCTC-3’) for the first round of PCR and primers E3 (5’-GTWATGCTYTGCATWCATGGCT-3’) and E4 (5’-AGCCGACGAAATCAATTCTGTC-3’) for the second round. The PCR cycling conditions for both rounds consisted of 35 cycles of denaturation for 30 seconds at 94°C, annealing for 30 seconds at 50°C, and extension for 40 seconds at 72°C. The PCR products were analyzed in a 1.5% agarose gel. The expected DNA band specific for the HEV was excised from the gel, purified with the AxyPrep DNA gel extraction kit (Axygen, USA) and cloned into a pMD T-vector (TaKaRa, Japan). Both strands of the inserted DNA amplicons were sequenced in a DNA analyzer (Applied Biosystems 3730 DNA Analyzer; Invitrogen, USA).
3.3. Phylogenetic Analysis
The viral nucleotide sequences were aligned using ClustaLW2.0. Phylogenetic trees were constructed using the neighbor-joining method and evaluated using the interior branch test method with MEGA software version 5.0. Prototype HEV strains used as references in the analysis and their GenBank accession numbers are shown in the phylogenetic tree.
4. Results
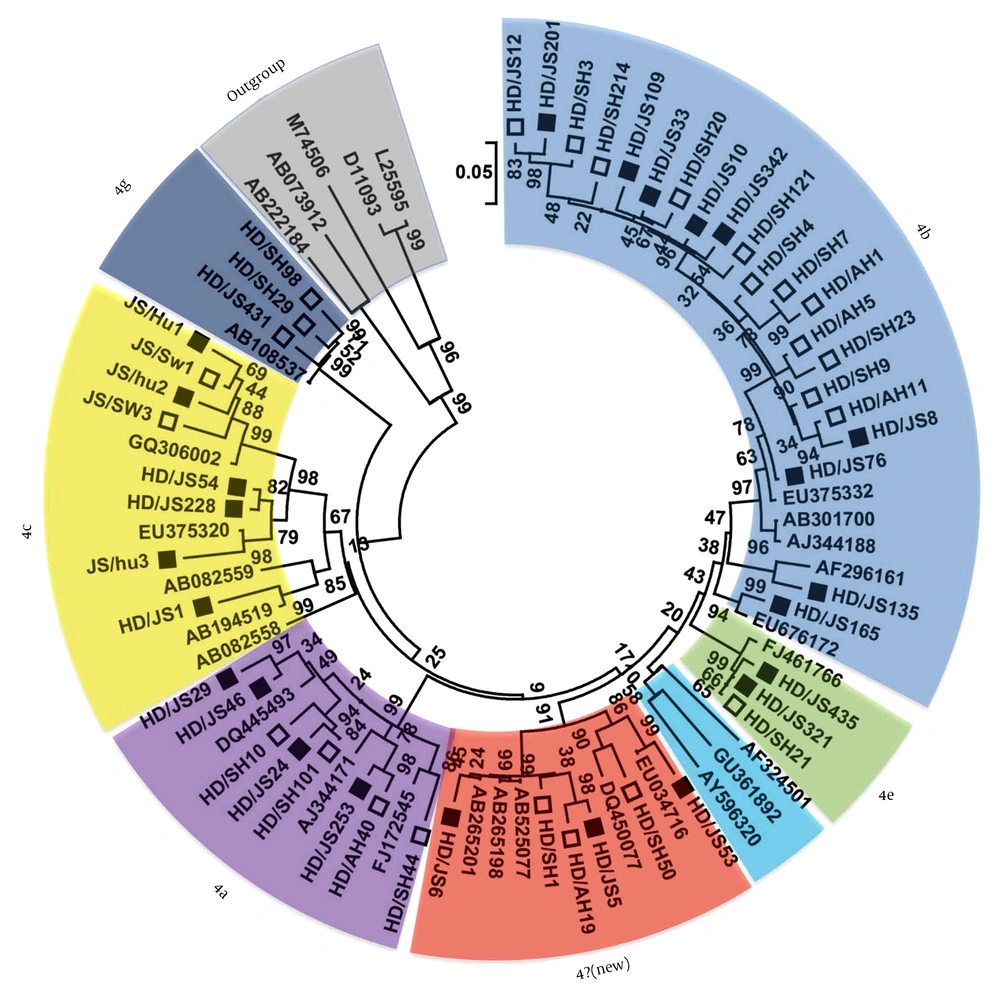
Our results indicated that 19.2% (24.125) of the serum samples were positive for HEV RNA, while 9.7% (28.290) of the swine fecal samples were positive for HEV RNA. Sequence analysis based on the PCR-amplified products (primer sequences were removed) indicated that the 52 HEV strains, including 24 from humans and 28 from pigs, all belonged to genotype 4 HEV, which suggested that genotype 4 was the main genotype prevalent in human and swine population in this area during our sample collecting time. The 52 genotype 4 HEV sequences shared more than 81.5% - 100% sequence identities and showed 49 distinct nucleotide sequences. In order to identify the subtype of the 49 genotype 4 HEV strains, phylogenetic analysis was performed based on the nucleotide sequences in the present study and other referenced HEV strains retrieved from GenBank and results were shown in Figure 1. Other 20 well-characterized subtypes of genotype 4 HEVs in a previous report (11) were also included in the phylogenetic analysis, where the GenBank accession were not marked with black or squares. Results indicated that the 49 genotype 4 HEV sequences in the present study were divided into 6 different subtypes, belonging to subtypes 4b, 4a, 4c, 4e, 4g and a new subtype, respectively.
The phylogenetic tree was produced with a 348 bp ORF2 sequence alignments of 49 strains in the present study and the reference sequences from GenBank, using the neighbor-joining method and evaluated using the interior branch test method with Mega 5 software. The percent bootstrap support is shown by values at the branch nodes of the tree. These values were the result of resampling the data 1000 times. The isolates identified in the current study were marked with black or squares. The white squares indicated the sequences from pigs and the black indicated the sequences from humans.
Twenty-one of the 49 (42.9%) genotype 4 strains were subtyped as 4b and shared 92.5% - 99.1% sequence similarity among themselves. The 21 subtype 4b strains included 12 strains from pigs and 9 strains from human populations. Eight strains in the present study felt into the cluster of subtype 4a, and shared 91.1% - 98.6% identities among themselves, including four strains from humans and four strains from pigs. The subtype 4c included eight strains from the present study, which shared 87.8% - 99.0% sequence identities among themselves, including six human originated and swine originated strains. Interestingly, four strains in this study could not be subtyped based on the defined subtypes, but closely clustered with several other strains in GenBank, forming a new subtype, sharing 92.2% - 95.4% sequence identities among themselves. Three strains including two strains from humans and one strain from pig clustered closely together and were subtyped into 4e. The last three strains were all from swine populations and typed into subtype 4 g.
5. Discussion
An increasing number of evidences indicate that HEV is enzootic and pigs were considered as the major reservoir for HEV infections in human populations (9, 13, 14). The zoonotic transmission of HEV from pigs to humans has been suggested all over the world. Genotype 4 HEV was the main cause of hepatitis E in China and involved in zoonotic transmission in several areas of China (13, 14). The HEV isolates identified from the same geographic region tended to cluster together (15, 16). However, recently several studies suggested the HEV strain from a certain limited region may have several different subtypes (9, 17). In the current study, we identified 49 genotype 4 HEV strains and phylogenetic analysis showed these isolates could be divided into six different subtypes, among which five subtypes contain isolates from both humans and pigs and the isolates from human and swine population closely clustered together, suggesting these five subtypes of genotype 4 HEV were involved in cross-infection between human and pigs in this area.
The first swine strain of HEV was isolated and characterized from a pig in the United States (2). Subsequently, many HEV samples from swine in over a dozen countries have been identified. In the present study, although we did not classify the swine fecal samples according to the pig age because of the label on part of the collection plastic bags were missing unexpectedly, the overall positive rate of HEV RNA (9.66%) was in agreement with several previously reports in other areas of China (13, 18). Hepatitis E virus isolates from human or pigs were divided into four distinct genotypes according to sequence and phylogenetic analyses. Recent several reports indicated that genotype 3 HEV was prevalent in swine and human populations in several regions of China (19, 20). However, the HEV strains in the present study all belonged to genotype 4, which may owe to the different sampling site and time.
In the present study, 42.9% (21.49) of the genotype 4 strains belonged to subtype 4b, including 12 strains from swine population and 9 strains from human, which suggested that subtype 4b is the main subtype prevalent in swine and human populations in this area. Phylogenetic analysis also revealed that the subtype 4b in the present study clustered with several other genotype 4 HEVs isolated from eastern China, while the other subtypes in this study clustered with HEVs from other regions in China or other countries, suggesting subtype 4b HEVs are indigenous subtype of eastern China while the other subtypes may not be indigenous to this area.