1. Background
Hepatitis B Virus (HBV) is the most common cause of liver infectious diseases in human, and the liver diseases related to this virus occur as acute and chronic hepatitis (1-3). Hepatitis B Virus is grouped into eight genotypes, from A to H, with distinctive geographical distribution worldwide. Among them, genotype D is predominant in the Mediterranean area and the Middle East (4, 5) and is reported as the most frequent genotype in Yemen (6), Egypt (7), Turkey (8) and Iran (4). On the contrary, genotypes A, B and C are dominant in Pakistan (9). Despite chemical and other infectious diseases, HBV is one of the most important causes of liver transplantation (LT) worldwide (10). LT offers the ultimate cure for patients with cirrhotic and hepatocellular carcinoma. However, in the absence of effective prophylaxis, the recurrence of hepatitis B after transplantation was as high as 75% (11). Also, enhanced viral replication resulting from immunosuppression and direct stimulatory effects of steroid therapy on the glucocorticoid-responsive enhancer region of the HBV genome can cause HBV reinfection after LT (12).
It is proved that HBV genotype is associated with certain prognoses, the clinical picture, and especially the outcome of response to treatment (13). Less than 1% of individuals with acute infection die and chronic infection is acquired by 5% - 15% of adults and 85% - 95% of newborns (14). The acute and asymptomatic forms of the disease are not treatable, however, complete and sustained suppression of viral replication remains the most important goal to treat patients with chronic HBV infection (15). The most common treatment of these patients involves oral administration of nucleoside/tide analogues such as LAM and adefovir (15). Therefore, treatment of chronic HBV infection with the nucleoside analogue and reverse transcriptase inhibitor, and lamivudine (LAM) are very effective in suppressing HBV replication without major side effects (16). LAM is a potent nucleoside analog Reverse Transcriptase Inhibitor (nRTI) used to treat chronic hepatitis B (17, 18). Also, combined prophylaxis of nucleotide analogues, and hepatitis B immune globulin (HBIG) significantly decrease the HBV recurrence and improve the prognosis of HBV infection in patients undergoing transplantation (10, 19, 20). However, one of the most important disadvantage of nucleoside/tide analogues is that resistant HBV mutants develop during long-term treatment, which can diminish the efficacy of these analogues in suppressing viral replication (15). It is reported that the cumulative rates of LAM-resistant HBV variants detection after one and two years of treatment were 10% and 56%, respectively (21). Also, hepatitis caused by mutations in the C domain of the HBV emerges in approximately 70% of the patients receiving LAM therapy for more than four years (15).
2. Objectives
The current study aimed to determine YMDD mutations, genotype identification, the rate and pattern of LAM resistance mutations in patients with liver transplantation (LT) receiving LAM therapy.
3. Patients and Methods
The study protocol was approved by the local ethics committee in the Department of Medical Ethics, Shiraz University of Medical Sciences, Shiraz, Iran and all participants gave informed written consent. A total of 1500 subjects were monitored at the Transplantation Center of Nemazee Hospital affiliated to Shiraz University of Medical Sciences, Shiraz, Iran from Jan 2013 to Jan 2014. Totally, 30 subjects who had received liver transplantation due to HBV infection and were HBV DNA negative (level under detection limit of 50 IU/mL) before transplantation were enrolled in a cross sectional study. These subjects were introduced to Transplant Research Center because of abnormal high levels of liver enzymes. All subjects had received a combination of 100 mg LAM daily plus low doses of HBIG at least for one year. Also, they were HCV/HIV RNA negative and received intense immunosuppression with tacrolimus plus mycophenolic acid to prevent rejection of liver transplant. The current study exclusion criteria were receiving LAM/HBIG irregularly or less than one year, and also coinfection with HCV and HIV. At the time of sampling for virology evaluation, blood coagulation tests including PT/PTT/INR in addition to regular routine liver function tests such as serum levels of albumin, AST, ALT, ALP and bilirubin (total/direct) were performed by the Central Laboratory of Nemazee Hospital, Shiraz, Iran.
3.1. Virological Assessment
Aseptic blood samples were collected from each patient and transferred on ice to the Laboratory of Gastroenterohepatology Research Center. The presence of HBsAg was determined by commercial enzyme immunoassay kit (Dia. Pro, Italy). For virus detection, HBV genomic DNA was extracted from serum samples by viral DNA extraction kit (Invitek, Germany). The presence/recurrence of HBV genome DNA in serum samples was evaluated by Polymerase Chain Reaction using two methods: HBV PCR detection kit (CinnaGen, Iran) and in-house gap-PCR method (13, 22). In the second method, there was a possibility to discriminate genotype D from non-D. To quantify virus genome, QPCR assay was also carried out using TaqMan-based commercial available kit (Amplisense Co., Russia) based on the manufacturer’s instructions. The method was performed by iQ5 Real-Time PCR detection system (Bio-Rad, America).
3.2. Amplification of Polymerase, Genotyping and Phylogenetic Analysis
To determine genotype, first a gap-PCR method was performed to differentiate genotype D from non-D (13, 22). Furthermore, to investigate the YMDD profile pattern in viruses, a new developed Nested-PCR for HBV polymerase (P) gene was employed. The functional part of P gene, that codes 80th - 240th amino acids of RT-polymerase and relevant overlapping region of HBs protein, was amplified using Nested-PCR. To amplify gene sequence and overlapping S gene, the external and internal primers were designed based on reference sequences according to Table 1.
| Primers | Sequences (5’-3’) | Function |
|---|---|---|
| YMF1 | TCACAATACCGCAGAGTCTAGAC | target amplification in the first cycle |
| YMR1 | AAATCCCAAAAGACCCACAAT | target amplification in the first cycle |
| YMF2 | CTCCAATCACTCACCAAC | P gene amplification for sequencing |
| YMR2 | GGGTTTAAATGTATACCCA | P gene amplification for sequencing |
All necessary precautions were observed to prevent cross-contamination and negative controls were included in different runs. Amplification was performed for the 25th and the 35th cycles of PCR and annealing temperatures of 55°C and 60°C, respectively. Amplified PCR products were purified by fragment DNA purification kit (Bioneer, South Korea) according to the manufacturer’s instructions. Purified HBV DNA was directly sequenced using ABI PRISMR 3700 DNA analyzer automated sequencer (Applied Biosystem, USA) by Genomics Service of Bioneer, South Korea.
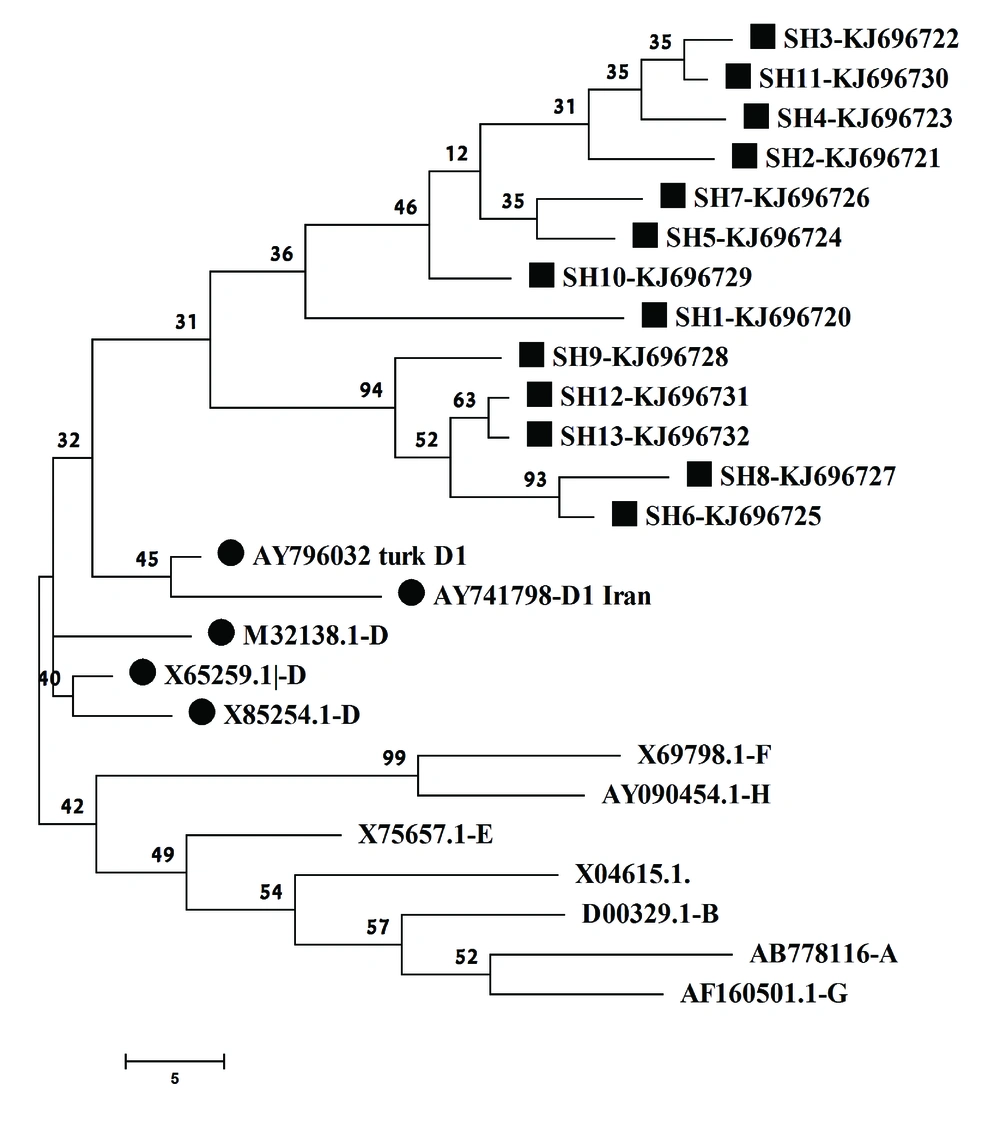
To perform the phylogenetic analysis and extrapolate point mutation patterns among RT domains, the HBV genome sequences were compared with those of defined HBV genotypes retrieved from GenBank. The selected accession number and relevant genotypes were: AB778116 (A), D00329.1 (B), X04615 (C), X75657 (E), X69798 (F), AF160501 (G), AY090454 (H), M32138 (D), X85254 (D), X65259 (D), AY741798 (D1 from Iran) and AY796032 (D1 from turkey). The selected sequences were from all HBV genotypes, while at least two D1 subgenotypes were also included to highlight the difference in phylogenetic analysis. The sequences were aligned using Clustal X to explore possible mutations. Besides simple eyes exploring through sequences, all new sequences were submitted to HBV-Resistance Interpretation Software from the Stanford HIV-DB (http://hivdb.stanford.edu). This software predicts the resistance against HBV drugs and expresses HBsAg-escape mutations based on the published data.
For aligned sequences, genetic distance was calculated using the maximum parsimony method algorithm due to high homology among the extracted sequences. The phylogenetic tree was constructed by the maximum parsimony and bootstrap resampling, and reconstruction was carried out 500 times to confirm the reliability of the phylogenetic tree. In the current study, Mega4 software was utilized for phylogenetic and evolutionary analysis.
3.3. Statistical Analysis
To explore any relationships between YMDD profile and clinical findings, the clinical data were analyzed by Chi-squared test and independent sample t-test using SPSS statistical package version 20 (SPSS Ltd., Woking, Surrey, UK) for Windows. Significant differences were considered when P < 0.05.
4. Results
4.1. Virological and Biochemical Evaluations
Totally, 13 out of 30 patients (43%) were recognized as HBV DNA positive. It was recognized after electrophoresis of PCR products and appearance of < 100 bp and approximately 220 bp bands in the gap-PCR and commercial methods, respectively. The clinical evaluations of the subjects were presented in Table 2 and clinical analyses comparison between HBV positive and negative subjects were shown in Table 3. These finding indicated no significant differences between HBV positive and negative subjects in all clinically measured parameters (P > 0.05) except PTT, total and direct bilirubin (P < 0.05).
| ID | HBV b | PT, s | PTT, s | INR | Bilirubin, μmol/L | AST, U/L | ALT, U/L | ALP, U/L | Alb, U/L | |
|---|---|---|---|---|---|---|---|---|---|---|
| T | D | |||||||||
| SH1 | 3+ | 12 | 27 | 1 | 1 | 0.3 | 76 | 71 | 393 | 4.3 |
| SH2 | 1+ | 11 | 13 | 1 | 0.9 | 0.1 | 18 | 21 | 192 | 4.5 |
| SH3 | 0.5+ | 13 | 15 | 1 | 1.3 | 0.4 | 11 | 22 | 244 | 4.6 |
| SH4 | 1+ | 12 | 15 | 1 | ND | ND | 14 | 37 | 127 | 4.4 |
| SH5 | 0.5+ | 13 | 36 | 1 | 0.8 | 0.3 | 21 | 31 | 135 | 5.7 |
| SH6 | 3+ | 16 | 40 | 1.4 | 0.9 | 0.3 | 218 | 258 | 284 | 4.5 |
| SH7 | 0.5+ | 17 | 33 | 1.5 | 1.3 | 0.6 | 24 | 19 | 162 | 4.9 |
| SH8 | 0.5+ | 13.5 | 29 | 1.2 | 1.9 | 0.4 | 24 | 28 | 349 | 4.7 |
| SH9 | 0.5+ | 14 | 20 | 1 | 0.8 | 0.3 | 38 | 40 | 273 | 5 |
| SH10 | 1+ | 16 | 38 | 1.4 | 2.6 | 0.3 | 30 | 29 | 117 | 4.9 |
| SH11 | Weak | 13 | 32 | 1 | 0.7 | 0.2 | 12 | 11 | 262 | 4.9 |
| SH12 | 0.5+ | 13 | 16 | 1 | 0.4 | 0.1 | 18 | 20 | 198 | 4.4 |
| SH13 | 0.5+ | 12 | 38 | 1 | 0.9 | 0.2 | 18 | 23 | 130 | 4.7 |
a Abbreviations: Alb, albumin; ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; D, direct bilirubin; INR, international normalized ratio; ND, not determined; PT, prothrombin time; PTT, partial thromboplastin time; T, total bilirubin.
b A scale of 0.5 - 3 determined for each positive electrophoresis result.
| Factors | HBV Status | P Value | |
|---|---|---|---|
| Positive | Negative | ||
| PT, s | 13.50 ± 0.50 | 13.80 ± 0.54 | 0.688 |
| PTT, s | 27.00 ± 2.81 | 35.40 ± 2.28 | 0.038 |
| INR | 1.11 ± 0.05 | 1.25 ± 0.09 | 0.206 |
| Bilirubin, μmol/L | |||
| Total | 1.12 ± 0.17 | 0.29 ± 0.04 | < 0.001 |
| Direct | 0.29 ± 0.04 | 0.93 ± 0.11 | < 0.001 |
| AST, U/L | 40.15 ± 15.55 | 21.14 ± 2.83 | 0.251 |
| ALT, U/L | 46.92 ± 18.06 | 22.36 ± 3.98 | 0.181 |
| ALP, U/L | 220.46 ± 24.68 | 193.57 ± 19.13 | 0.394 |
| Alb, U/L | 4.73 ± 0.10 | 4.73 ± 0.06 | 0.983 |
a Abbreviations: Alb, albumin; ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; D, direct bilirubin; INR, international normalized ratio; PT, prothrombin time; PTT, partial thromboplastin time; T, total bilirubin.
4.2. Drug Resistance Mutations Analysis
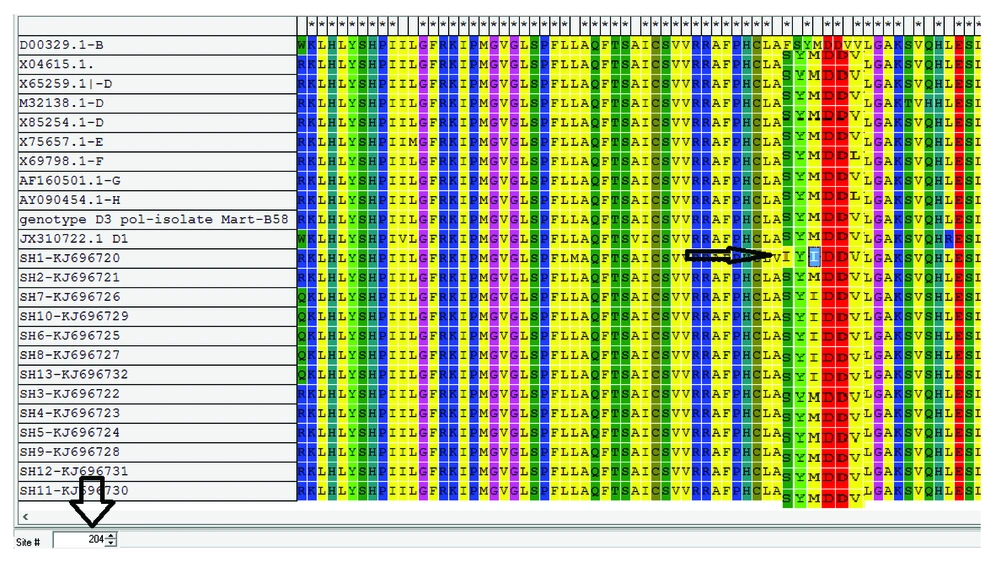
The YMDD motif, harboring sequence of P gene in all these 13 subjects, were amplified and sequenced. A 510 bp band covering amino acid sequences 80 - 240, amplified by Nested-PCR, contained the major part of P and middle part of S genes. The final gene sequences of this study were deposited in GenBank under accession numbers of KJ696720-KJ686732. After mutation analysis of aligned sequences by bioinformatics software, four major types of point mutations related to drugs resistance were detected in 6 out of 13 subjects (46.1%) as demonstrated in Table 4. As expected, in all the 6 subjects, M204I point mutation, responsible for complete resistance to LAM or telbivudine (TEB) and intermediate resistance to entecavir (ENC), was detected as dominant mutation. Also, according to P sequences, there were no evidences of resistance mutations to tenofovir and adefovir. However, it should be noticed that natural resistance to these drugs is scarce albeit following treatment is possible. As a rare case, natural mutation of S202I, responsible for resistance to ENC, occurred before starting drug consumption (in patient SH1). This type of mutation has a clinical significance because ENC is accepted as the first line drug of choice for HBV treatment in subjects receiving transplant. Only in one of the subjects (designated as SH1), three important mutations coexisted, which were responsible for viral resistance to LAM, ENC and TEB (Figure 1).
| ID | DR | HBsAg | Viral Load | Mutation, s | EM |
|---|---|---|---|---|---|
| SH1 | lamivudine entecavir telbivudine | ND | 1.7 × 106 IU/mL | A200V L180M M204I S202I | Y134N |
| SH2 | NDR | ND | 8.0 × 103 IU/mL | NM | NEM |
| SH3 | NDR | + | 7.5 × 104 IU/mL | NM | NEM |
| SH4 | NDR | ND | 2.5 × 104 IU/mL | NM | NEM |
| SH5 | NDR | ND | 3.5 × 103 IU/mL | NM | NEM |
| SH6 | lamivudine telbivudine | 3+ | 1.3 × 106 IU/mL | M240I | I110V G145R |
| SH7 | NDR | + | 3.1 × 104 IU.mL-1 | NM | NEM |
| SH8 | lamivudine telbivudine | ND | 2.0 × 105 IU/mL | M240I | NEM |
| SH9 | lamivudine telbivudine | ND | 9.7 × 104 IU/mL | M240I | G145R |
| SH10 | NDR | ND | 6.0 × 104 IU/mL | NM | NEM |
| SH11 | NDR | + | < 50 IU/mL | NM | |
| SH12 | lamivudine telbivudine | ND | 1.2 × 105 IU/mL | M240I | NEM |
| SH13 | lamivudine telbivudine | ND | 5.3 × 104 IU/mL | M240I | I110V T127P G145R |
a Abbreviations: EM, escape mutation; DR, drug resistance; ND, non-detectable; NDR, no drug resistance; NEM, no escape mutation; NM, no mutation.
4.3. Escape Mutations Analysis
Besides vaccine escape mutations that occur in S gene and cause evading of virus from neutralizing antibodies were also detected in the current study. The obtained results demonstrated three types of escape mutations (Y134N, I110V, G145R) in four subjects with drug resistance mutations and higher viral load (Table 4) as well. The level of HBsAg was also measured by commercial ELISA assay. The analysis indicated no positive correlation between drug resistances related mutations and levels of HBsAg; moreover, detectable amounts of HBsAg existed only in the four subjects. It may be due to passive HBIG application that interferes with HBsAg assay. Therefore, in the subjects with detectable HBsAg it could be concluded that immunoglobulin treatment failed or partially devastated.
4.4. Biochemical-Virological Relationships
As a key virological element to predict viral response to treatment, HBV viral load was performed by commercial kit. The results demonstrated that the 13 subjects can be classified into two groups: ≥ 105 (four patients) and < 105 IU/mL (nine patients). It is clearly demonstrated that in the subjects with no drug resistance related mutations, the viral load is lower than 105 IU/mL. On the other hand, the viral count in 83% of subjects with drug resistance (five out of six) was higher than that of subjects with drug susceptibility, which means despite HBV recurrence, LAM plus HBIG can control the viral load below the effective limit (< 100000 IU.mL-1). Between these two groups, significant differences were observed in serum AST, ALT and ALP levels (P < 0.001, P = 0.001 and P = 0.034, respectively). Also, the analysis demonstrated significant positive correlation between viral load and serum levels of AST (R2 = 0.577, P = 0.003), ALT (R2 = 0.495, P = 0.007), and ALP (R2 = 0.442, P = 0.013). Further clinical analysis between mutant and normal YMDD patients indicated that although patients with normal YMDD, compared to mutant ones, had higher values of PT, INR, total and direct bilirubin, albumin and lower levels of PTT, AST, ALT and ALP, the differences were not statistically significant (P > 0.05).
4.5. Sequencing and Phylogenetic Analysis
The phylogenetic analysis confirmed that the virus isolates were clustered in the genotype D branch with other genotype D HBV reference genes (Figure 2). The phylogenetic analysis data were completely in agreement with those of the gap-PCR results, predictable for the research area. Although the sequence was not suitable for subgenotype determination, S gene sequence analysis demonstrated that all 13 subjects belonged to subgenotype D1 and D3 based on drug resistance online database albeit the data were not supported by phylogenetic analysis (data not shown).
5. Discussion
In the current study, the virological parameters and mutation analysis of YMDD motif were assessed among patients receiving LT due to HBV infection with more than one year of LAM/HBIG treatment. LAM treatment was selected because it is a safe and effective drug to treat recipients with recurrent hepatitis B in a short time (10). The study findings showed HBV recurrence among 13 out of 30 subjects, consistent with the previous reports (23-25). However, the number of subjects with HBV after LT was much less than that of the study by Perrillo et al. They reported that 45 out of 52 patients (86%) with chronic hepatitis B after liver transplantation had detectable HBV DNA in serum (26). The data also indicated some major mutations regarding LAM and TEB resistance. Currently, antiviral drug resistance is a major problem in the management of patients with chronic HBV infection and LAM resistance may be due to YMDD mutations as demonstrated in the current study and reported previously (27). LAM inhibits viral DNA polymerase in HBV, and its related resistance among subjects with HBV infection during the first year of treatment is reported as 10% - 45% (24). The approval of LAM revolutionized the treatment of chronic hepatitis B and it was also evaluated in patients with recurrent hepatitis B after LT in the experimental studies (26-28). One of the most important drawbacks with LAM usage is drug-resistant HBV mutants selection (20) and these types of genotype resistance mutations were detected as levels of 32%, 38%, 49% and 66% after one, two, three, and four years of treatment, respectively (21, 23, 25). Although the number of patients in the current study was small, it showed that changes in the YMDD region were frequently observed both spontaneously and under LAM therapy. Owing to the low fidelity of the virus polymerase, the high replication rate and under the treatment pressure, different mutations occur throughout the HBV genome (29). The results of mutations were similar to those of the previous studies reporting that the most common mutation affects the highly conserved YMDD motif, resulting in a methionine to valine or isoleucine substitution at codon 204 (M204V/I) and between them, M204I mutant is significantly more resistant to LAM than the M204V mutant (27, 30). Also, the frequency of YMDD mutation in the current study subjects (46%) was approximately higher than those of the previously reported values of 27% (26) and 21% (31).
In addition, the current study clinical findings, in line with the other previous reports (23, 32, 33), indicated that higher viral load and existence of LAM resistance mutations had significant correlation with higher liver enzyme levels. However, the levels of liver enzymes and other related biochemical analyses cannot be insisted on as definitive diagnostic tools. Moreover, viral titer of mutated and normal YMDD motif patients indicated that in YIDD mutation, the viral load were higher than those of the wild type YMDD presented elsewhere. The sequential analyses of the current study genes also revealed the natural presence of ENC resistance mutation (S202I), not previously reported in Iran. Since this mutation was detected in a small population it should be considered in future studies. LAM is a valuable drug in transplantation and as well as for subjects with HBV relapse; therefore, the natural mutations should be investigated more in a bigger population. The limitations of the study include the relatively small number of subjects, the lack of a randomized control group and no liver histological evaluations. Since the current study did not assess liver histology in the subjects who harbored these viral mutants, it is difficult to be sure if the modest change in biochemical parameters underestimated the amount of damage to the liver.
Sequence analysis by drug resistance database demonstrated that all HBV strains mostly belonged to the subgenotype D1 and D3, but it was not supported by tree construction, since amplified sequence was not able to discriminate between subgenotypes. Therefore, for the next step it suggested that another part of genome such as core and pre-S parts submitted for sequencing. Different genotypes of HBV possess distinct geographical distribution and among them, genotype D is the predominant strain in the Middle East (34). The current study findings about HBV genotypes were in line with the previous reports which demonstrated that genotype D of HBV is highly predominant in Iran (35, 36). Antiviral drug response between HBV genotypes is different and it is reported that genotype D of HBV has a lower response rate to LAM compared to the other genotypes, especially genotype A (37). The study found M204I as the dominant mutation in all LAM resistant patients. These findings, consistent with previous studies (38-40), demonstrated that the most common mutation affects the highly conserved motif in the catalytic domain of the HBV P gene is M204I. Also, the virological relapse rates, genotypic resistance to LAM, was detected in 6 (46.1%) subjects after at least one year of LAM treatment that is approximately similar to those of the previous report (24). The current study detected no or low levels of HBsAg in the subjects undergoing LT and there was no correlation between the existence and type of antiviral drug resistance mutation and HBsAg level. It is reported that the rate of HBsAg seronegativity after 8 years of treatment with LAM in subjects undergoing LT was 88% (41). A logical positive correlation between duration of LAM therapy and percentage of HBsAg seronegativity was reported in the literature. It means that by increasing the time of LAM therapy, the rate of HbsAg seronegativity in LT patients also increases. However, in the current study with one year of LAM treatment, the rate of HBsAg seronegativity was approximately high (69%). This may be due to the simultaneous use of HBIG with LAM that can be interfered with Ag-Ab interaction and decreases the levels of HBsAg detection (29).
In conclusion, passive immunization of subjects with HBV infection and HBIG, treatment with nucleotide analogues, and possibly failed active vaccination of HBV infected individuals (similar to the current study due to existence of escape mutation) can favor the outgrowth of mutants (42, 43). However, if the current study results are confirmed in larger studies, a subgroup of subjects undergoing liver transplanted with YMDD mutations could benefit from stopping the prolonged LAM therapy and replacing other effective nucleoside/tide analogs.