1. Background
Chronic hepatitis B (CHB) continues to be a major global health problem, despite widespread vaccination and improving therapeutic options, which currently include both immunomodulatory and antiviral interventions. Hepatitis B surface antigen (HBsAg) loss and anti-HBs seroconversion have been observed in a minority of patients during antiviral treatments, or after its interruption (1, 2). Treatment interruption has been indicated as a potential strategy for time-limited antiviral treatment (3), while other innovative antiviral/immunomodulant integrated strategies have been proposed for a functional cure (4, 5).
In view of the prospective increase in the therapeutic options, there is an urgent need for improved and reliable noninvasive virological markers to identify patients who might benefit from any of these strategies. For instance, the dynamics of the serum HBsAg in the treated patients, as a surrogate marker of liver colonization, has proven to be the most reliable predictor of clinical success in patients treated with interferon (6), where HBsAg decay was found to correlate intrahepatically with the decay of covalently closed circular DNA (cccDNA). However, this parameter is a poor predictor of a viral load reduction (measured either before or during treatment) and an intrahepatic cccDNA burden (7, 8). cccDNA is undoubtedly the best marker of hepatic colonization, and a more accurate comprehension of the relationship between intrahepatic and peripheral virological markers is needed before we can use the latter as predictors of successful antiviral interruption, and spare needle biopsies.
cccDNA is formed after the cellular processing of replicative HBV DNA, and persists in the nucleus of the infected cell as a stable, histone complexed minichromosome. Its intranuclear pool increases by processing de novo reverse transcribed templates to a variable number of copies until an equilibrium is reached, usually 10 - 100 copies per infected cell (9, 10). In addition to the cccDNA, the viral DNA pool in the infected hepatocytes is represented by molecules derived from the activity of the reverse transcriptase (RT), replicative intermediates in various stages of completion, up to the encapsidated relaxed circular forms of viral genomes (rcDNA). These molecules are normally much more abundant than cccDNA, but are significantly reduced in patients treated with RT inhibitors (11-13). The ratio between the total DNA (tDNA) and cccDNA in the treated patients with undetectable viral loads may, therefore, represent a useful marker of residual viral replication and colonization.
However, since bioptic sampling is an invasive procedure, and seldom indicated in patients on antiviral treatment, a study addressing the dynamic relationship between the intrahepatic and peripheral parameters is needed for revisiting the use of the peripheral parameters as predictors of a successful treatment interruption. Among the peripheral parameters, cccDNA in the leukocytes may also provide additional information for this goal. In fact, as it has been previously demonstrated in many different studies, peripheral blood lymphocytes are hosts to viral replication (14-17), while granulocytes may be passively carrying the different forms of HBV DNA as a result of their phagocytic activity on dead hepatocytes in the liver.
2. Objectives
The aim of this study was to develop versatile and reliable real-time PCR methods for the quantitative detection of HBV tDNA and cccDNA, and to explore their use in clinical samples from HBV infected, treated, or untreated patients.
Particular attention was dedicated to simplifying the procedure for the differential amplification of cccDNA, in order to overcome the need for using DNAse. This enzyme has been used in previous studies to degrade single strand portions of the relaxed forms of HBV DNA that may prime the PCR reaction leading to false-positive results (18, 19), but is unpractical for application in a clinical laboratory routine: other studies have described DNAse free assays (20-23).
3. Patients and Methods
3.1. Patients and Clinical Samples
Biological samples were collected from HBV-positive patients with chronic HBV infections attending the outpatient clinic at the national institute for infectious disease “L. Spallanzani” in Rome, between 2012 and 2013, with the exclusion of HIV-1 positive patients. Local ethics committee approval was obtained, and each patient provided written consent to the research use of his/her biological samples. In particular, in order to test the new real-time PCR, 98 sequential fresh blood samples from those patients attending the clinic for routine HBV viremia testing were collected (31 untreated and 67 treated patients). Liver tissue samples and peripheral blood samples were collected from 6 untreated and 6 treated patients (9 from fine needle liver biopsies and 3 from liver resections), collected for the purpose of diagnostic histology.
3.2. Plasma HBV DNA Quantitation
The plasma total HBV DNA was measured using the COBAS AmpliPrep/COBAS TaqMan HBV test (Roche Molecular Diagnostics, Pleasanton, CA, USA).
3.3. Serological Testing
All of the serological testing was performed using the carbonylmetalloimmunoassay (CMIA) technique on the Architect platform (Abbott Diagnostics, Wiesbaden, Germany). The system provides standardized quantitative results for the HBsAg and anti-HBsAg, expressed as international units and milliInternational units (IU/mL and mIU/mL), respectively.
3.4. Isolation of Peripheral Blood Mononuclear Cells from Whole Blood
Peripheral blood mononuclear cells (PBMC) were isolated from EDTA blood by Ficoll-Paque (Pharmacia). In particular, 3 mL of Ficoll were added in a centrifuge tube, and 6 mL of diluted blood (with sterile PBS 1:1) were carefully layered onto the Ficoll solution. After centrifugation at 600 RCF for 20 minutes, the upper layer, which contained the plasma, was saved for later use, and the ring with the PBMC was harvested, washed twice with PBS, and pelleted. The granulocytes were harvested from the buffy coat remaining at the bottom of the Ficoll gradient. In addition, the erythrocytes were removed by lysis using NH4Cl (8 g/L), and the remaining leukocytes were washed twice and pelleted. The pellets of the PBMC and the granulocytes were directly frozen and stored at −80°C until analysis.
3.5. Sucrose Density Gradient Preparation
A continuous sucrose density gradient was obtained with a gradient-maker (Bio-Rad, Hercules, CA, USA) using 65% and 15% sucrose solutions, and centrifuged in a swinging bucket rotor (TST41.14, Kontron, presently Sorvall, Thermo Fisher Scientific Waltham, MA, USA) at 120000 RCF on an Optima L-90K ultracentrifuge (Beckman Coulter, Indianapolis IN, USA) for 5 hours.
3.6. DNA Extraction
The total DNA was extracted from needle liver biopsies (0.5 - 1.0 cm), from cell pellets, and from plasma samples by a QIAsymphony DSP virus/pathogen Midi Kit (Qiagen GmbH, Germany), using the automated QIAsymphony instrument (Qiagen).
3.7. Specific Primers and Probes for HBV cccDNA, tDNA, and hTERT Amplification
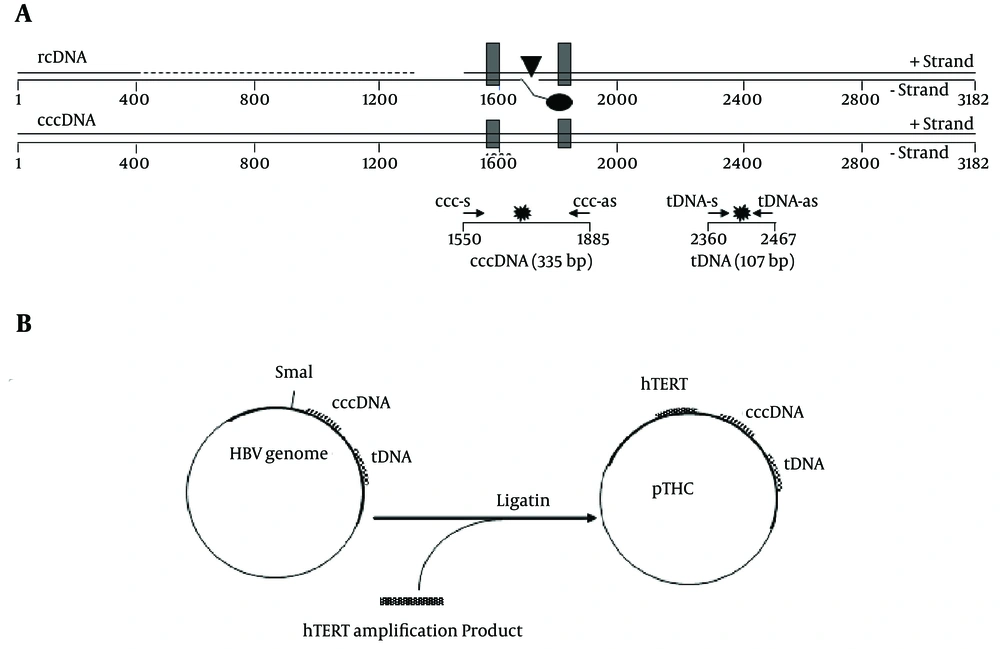
Relaxed circular HBV DNA (rcDNA) has a gap in the minus strand, near the DR1, and an incomplete plus strand with a defined 5' end, near the DR2. The cccDNA can, therefore, be preferentially amplified with primers flanking the gap region and the incomplete strand of DNA. Total HBV primers amplify a conserved region in the core gene and, therefore, detect all forms of HBV DNA, including the rcDNA and cccDNA. Specific primers and new probes were originally designed (although some are very similar to those already described in the literature for the molecular constraints of the peculiar genomic region of interest) to amplify the HBV cccDNA and HBV tDNA (Figure 1A and Table 1).
A, Schematic representation of the HBV genome, shown in a linearized form, according to the conventional numbering of the nucleotide positions of the HBV genome. The viral rcDNA is partially double-stranded. The direct repeats, DR1 and DR2, used in the HBV replication are shown as black boxes. The location of the basal core promoter (BCP) is indicated by a black triangle. The real-time PCR primers used for the cccDNA and tDNA detection (ccc-s/ccc-as and tDNA-s/tDNA-as, respectively) are shown as small arrows. The positions of the fluorescent probes (ccc-probes and tDNA-probes) are shown as stars; B, Schematic description of the construction and structure of multistandard plasmid pTHC.
| Primer | Location, nt a | Sequence (5' - 3') | Amplicon Length, bp |
|---|---|---|---|
| ccc-s | 1550 - 1569 | CGTCTGTGCCTTCTCATCTG | 335 |
| ccc-as | 1885 - 1865 | AAGGCACAGCTTGGAGGCTTG | 335 |
| ccc-probe | 1798 - 1774 | FAM-ACCAATTTATGCCTACAGCCTCCTA-BHQ1 | 335 |
| tDNA-s | 2360 - 2376 | AGGTCCCCTAGAAGAAG | 107 |
| tDNA-as | 2467 - 2446 | TGAGTCCAAGGAATACTAACAT | 107 |
| tDNA-probe | 2375 - 2396 | FAM-AGAACTCCCTCGCCTCGCAGAC-BHQ1 | 107 |
a Nucleotide positions refer to the prototype HE974382.1 (HBV genotype D4 complete genome, isolate Mart-B70).
To normalize the HBV tDNA and cccDNA content in the tissue extracts, the cellular content was determined using real-time PCR targeting the hTERTgene, as previously described (24).
3.8. HBV cccDNA, tDNA, and hTERT Quantitation by Real-Time PCR Assay
Real-Time PCR amplification was performed using the DyNAmo Flash Probe qPCR Kit (Thermo Scientific, Life Science Research), in a 30 µL reaction mixture containing 5 µL of extracted DNA or standard plasmid pTHC dilutions. The final concentrations of the probes and primers were 0.1 µM and 0.5 µM, respectively.
The thermal cycling conditions were as follows: 10 minute at 95°C, followed by 45 cycles of 20 s at 95°C and 30 s at 62°C to amplify the HBV cccDNA and hTERT; and 8 minute at 95°C, followed by 50 cycles of 10 s at 95°C and 50 s at 56°C to amplify the HBV tDNA. The amounts of the HBV tDNA, cccDNA, and hTERT in each sample were determined by extrapolation from the external dilution curves of a multi-standard plasmid (pTHC) of known concentration, which incorporated all three templates in equimolar amounts. The results were expressed as the log10 HBV tDNA and cccDNA copies/106 cells, respectively.
3.9. Statistical Analyses
Statistical analyses were performed using GraphPad Prism 4.02, and correlations between two variables were tested by using the Pearson’s analysis. Moreover, the quantitative parameters were evaluated in different populations by using the Mann-Whitney non-parametric test.
4. Results
4.1. Generation of the Multistandard Plasmid (pTHC)
The multistandard plasmid for the quantitation of the tDNA, cccDNA, and hTERT was constructed by inserting the hTERT target sequence in an HBV molecular clone originally designed and constructed for reverse transcriptase phenotyping. This molecular clone contained the whole HBV genome, with a deletion in the RT sequence, substituted by a SmaI site for the insertion of exogenous RT sequences. The hTERT amplification product, obtained by using a proofreading polymerase (PWO, Roche) was inserted in this site in place of the RT sequence, and the resulting plasmid, containing all 3 target sequences described in the methods section, was named pTHC (Figure 1B).
This pTHC construct was used to transform competent Escherichia coli cells using the One Shot TOP10 system (Invitrogen Life Technologies). A clone containing the expected sequence was selected, and the plasmid was extracted with a QIAprep Miniprep kit (Qiagen) and stored at –80°C in aliquots. The plasmid concentration was determined by spectrophotometry at 260 nm, and for each run of the real-time PCR, a standard curve was plotted, from 103 to 106 copies/reaction, by diluting an aliquot of the pTHC.
4.2. Validation of HBV tDNA Quantification Real-Time PCR Assay with Reference to COBAS Assay
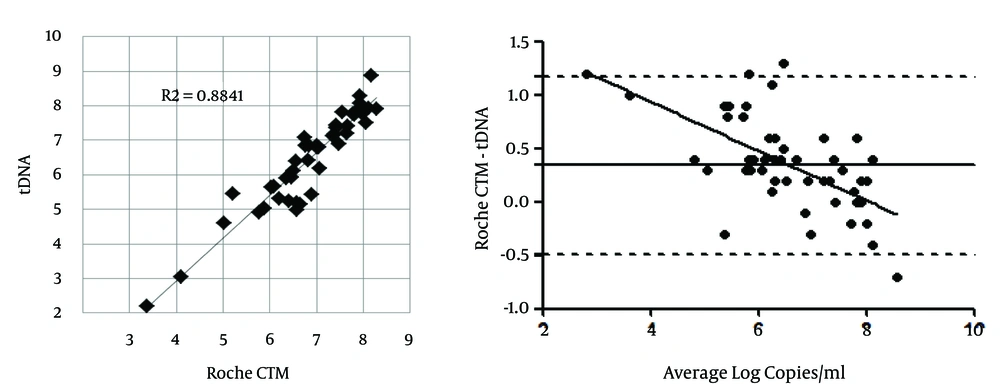
The HBV tDNA real-time PCR assay was evaluated by comparing the results obtained from the plasma of CHB patients with those obtained with a commercial diagnostic assay, the COBAS AmpliPrep/COBAS TaqMan HBV Test (CTM). Sixty clinical plasma samples from consecutive patients infected with various HBV genotypes were tested with both assays (a survey from a routine resistance testing analysis from the same center showed the following genotype prevalence: genotype D = 73%, A = 20%, C = 2%, F = 1.7%, E = 1.2%, B = 1.2%, and others = 0.8%, unpublished data). The IUs were converted to copies by using the ROCHE conversion figure: 1 IU = 5.82 copies. The correlation between the results obtained with this real-time PCR method and the ROCHE CTM assay (on 45 samples < 170000000 IU/mL, the maximum quantified by the ROCHE assay) was evaluated by regression, and a good correlation was observed (R2 = 0.884; P < 0.0001) (Figure 2A). The remaining samples (15 samples > 170000000 IU/mL, i.e. > 989400000 copies/mL by the ROCHE assay) still quantified by the in-house tDNA assay (10 results > 989400000 copies/mL and 5 > 300000000 copies/mL by the latter test). The Bland-Altman analysis (Figure 2B) was performed to identify the quantification bias depending on the copy number; a slight quantification bias (0.3 log copies/mL) was apparent, mostly for low copy numbers, when compared to the ROCHE assay.
4.3. Sensitivity of the Hepatitis B Virus DNA Quantification the Real-time PCR Assays
The sensitivities of both real-time amplifications were determined using the serially diluted pTHC (Probit analysis). The lower limit of detection was 4.8 copies/reaction for the tDNA detection (which for plasma corresponds to 15.2 IU/mL, according to the extraction/elution volume and the ROCHE copy unit conversion Figure), and 13 copies/reaction for the cccDNA detection.
4.4. Specificity of the Hepatitis B Virus cccDNA Quantification Real-time PCR Assay
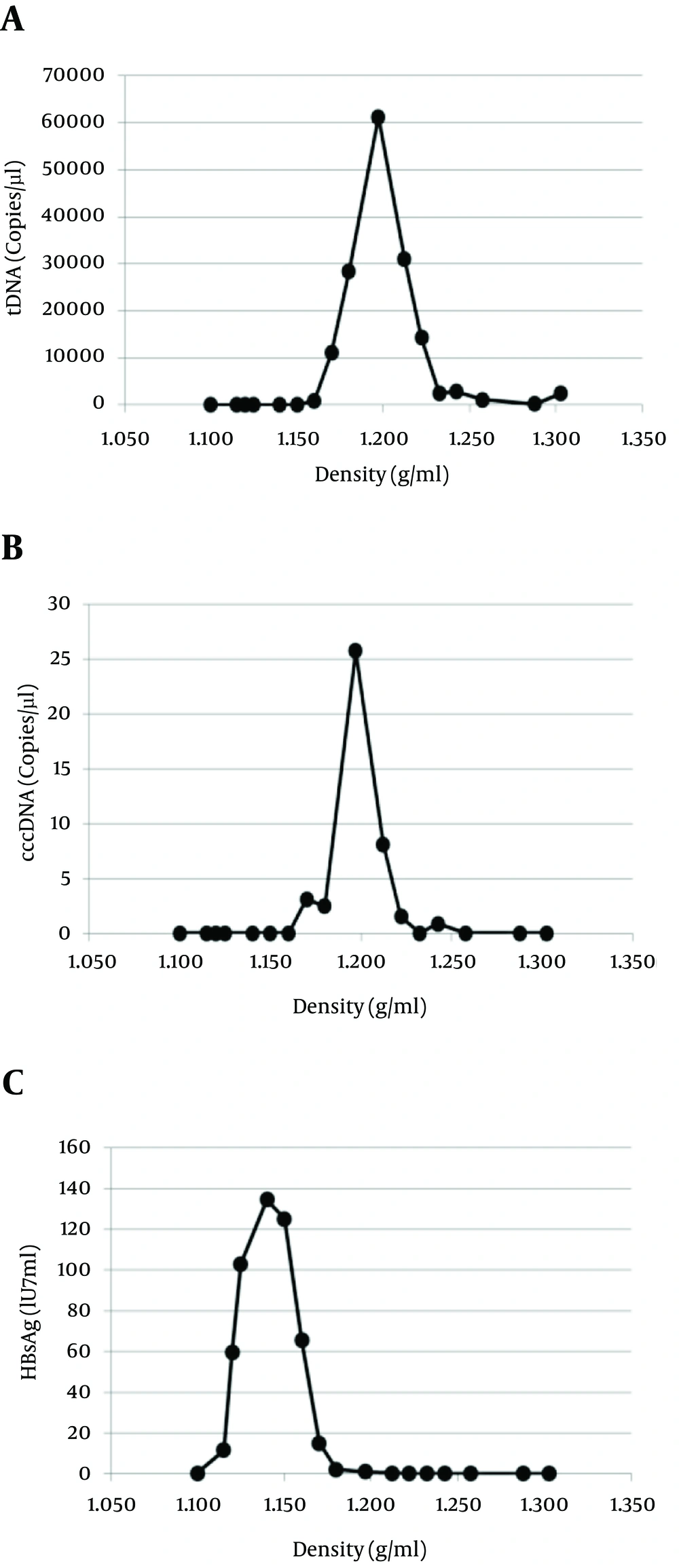
A critical issue for cccDNA assays is their ability to preferentially quantitate this molecule, without gross interference by the much more abundant other forms of HBV DNA. To test the specificity of the assay, HBV Dane particles were isolated from a plasma sample from a highly viremic HBV patient (108 IU/mL) by means of a sucrose density gradient. The cccDNA and tDNA were assessed in 17 gradient fractions. As shown in Figure 3A, a sharp peak in the concentration of the tDNA, corresponding to the Dane particles (complete virions), was reached in fraction 10 at a density of 1.197 g/mL: 61300 copies/µL. In the same fraction, the cccDNA reached a concentration of 25 copies/µL, with a ratio of 1/2452 to tDNA (Figure 3B). As expected, the bulk of the HBsAg was detected at lower densities (subviral particles, devoid of HBV cores); Dane particles account for a tiny minority of the HbsAg in plasma, and no HBV DNA was found to be associated with subviral particles (Figure 3C). Thus, the cccDNA assay demonstrated a sensitivity for the non-ccc templates, well below 10 - 3 when compared to the ccc templates. Since the ratios between the cccDNA and tDNA obtained from tissue and cell samples ranged from 1 to 1/100, the specificity of the described assay can be considered adequate for the reliable quantification of cccDNA in cellular samples.
4.5. Intrahepatic HBV DNA
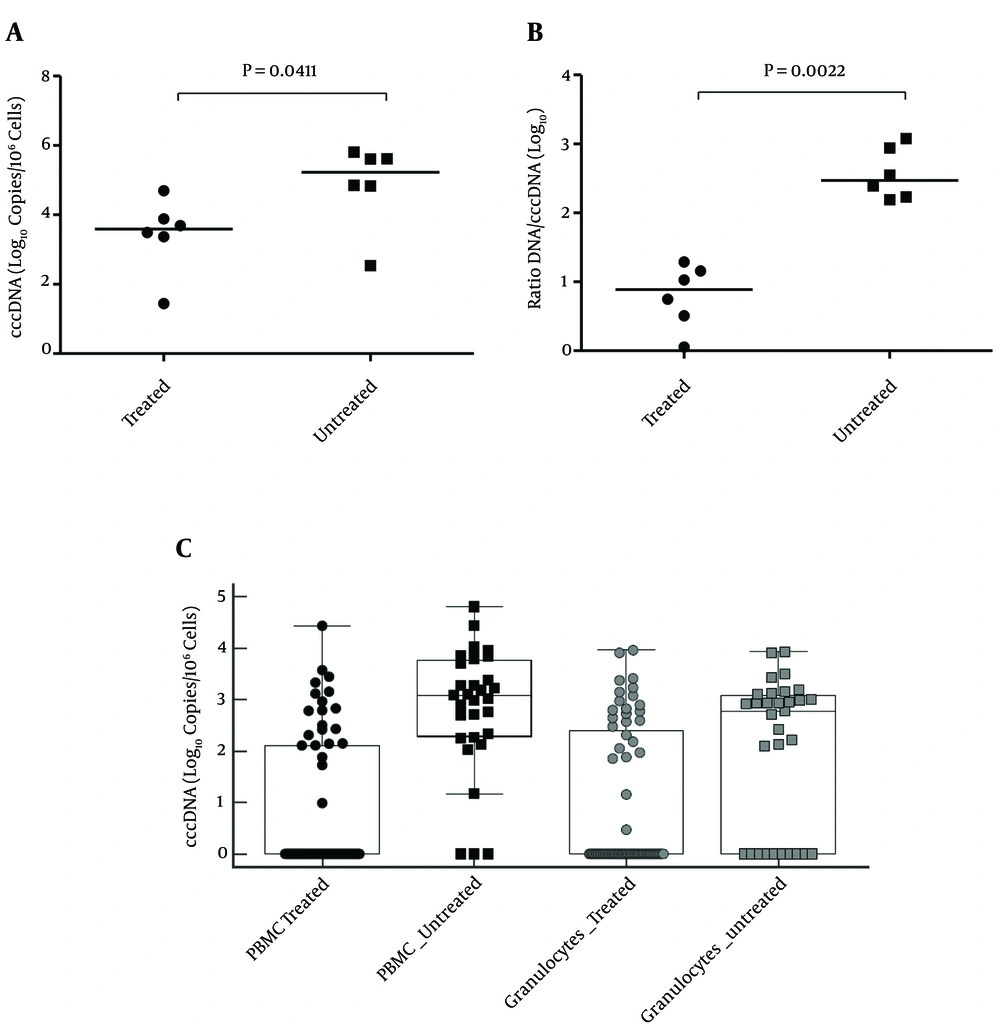
Intrahepatic HBV DNA was quantified for the comparison of 6 untreated and 6 long term-treated patients from fine needle liver biopsies (3 from liver resections for hepatocellular carcinoma). All of the treated patients had been treated for more than 2 years (median 4.6 years), and had undetectable or < 20 UI/mL HBV DNA in the plasma; the untreated patients were highly viremic. Table 2 shows the biochemical and virological data of the 2 groups of patients. In general, the treated patients were older, with more advanced liver histologies and bilirubin levels, but less altered transaminases and lower HbsAg. The tDNA amplification included both the relaxed and ccc forms of the HBV DNA. The total concentration of the cccDNA in the liver, a measure of the hepatic colonization by the virus, showed a 60-fold reduction in the treated patients (median 4010 vs. 241250 copies/106 cells, P = 0.0952) (Figure 4A). This is not overly impressive after years of viral suppression in the peripheral blood, suggesting that antiviral treatment is relatively ineffective at reducing the burden of infected hepatocytes in the majority of patients. In addition, as a measure of the replicative capacity of the virus in the liver parenchyma of the patients, the direct ratio between the tDNA and cccDNA copies was calculated as the yield of the tDNA molecules per cccDNA molecule, and defined as the replicative index (RI). This ratio has its minimum at around 1, as in the complete absence of viral replication, and the tDNA assay detects only cccDNA molecules, yielding the same results as the cccDNA assay. As shown in Figure 4B, there is a clear-cut and highly significant difference between treated and untreated patients for this parameter. The treated patients showed a highly reduced RI compared to the untreated patients (median = 8.09 vs. 298.5, P = 0.0022), making this parameter the best possible marker of intracellular antiviral activity.
| Patient | Gender | Age a | Total Bilirubin | Direct Bilirubin | AST | ALT | HBsAg, IU/mL a | HBeAg | METAVIR Grade | Stage | Genotype |
|---|---|---|---|---|---|---|---|---|---|---|---|
| T1 | F | 64 | 0.4 | 0.2 | 52 | 86 | 238 | negative | A1 | F2 | D |
| T2 | M | 61 | 0.9 | 0.3 | 25 | 25 | 138 | negative | A1 | F3 | D |
| T3 | M | 76 | 0.7 | 0.2 | 16 | 16 | 90 | negative | A1 | F3 | D |
| T4 | F | 43 | 3.9 | 3 | 91 | 84 | 2722 | positive | A1 | F4 | A |
| T5 | M | 51 | 0.4 | 0.2 | 28 | 36 | 1439 | negative | A1 | F4 | D |
| T5 | M | 56 | 3 | 2.5 | 95 | 127 | 4997 | negative | A2 | F4 | D |
| U1 | M | 44 | 2.2 | 1.6 | 138 | 259 | 912 | negative | A2 | F2 | D |
| U2 | F | 16 | 1 | 0.4 | 64 | 98 | 6854 | negative | A1 | F1 | D |
| U3 | M | 33 | 0.6 | 0.2 | 47 | 111 | 2102 | negative | A1 | F2 | D |
| U4 | M | 47 | 0.6 | 0.2 | 46 | 95 | 179298 | positive | A1 | F2 | F |
| U5 | M | 26 | 0.3 | 0.1 | 43 | 30 | 87280 | positive | A1 | F1 | E |
| U6 | M | 58 | 0.6 | 0.3 | 42 | 80 | 124925 | positive | A2 | F2 | A |
a Abbreviations: F, female; M, male; Tn, treated patients; Un, untreated patients.
b Parameters significantly different between the 2 groups.
A, intrahepatic cccDNA load in the liver tissue of treated and untreated patients; B, replicative index (RI) of HBV in the liver tissue of treated and untreated patients; C, HBV DNA content in peripheral blood leukocytes. Results are expressed as medians; boxes represent standard deviations. The 0 on the Y axis corresponds to “not detected”.
4.6. cccHBV DNA in Lymphocytes and Granulocytes
In order to investigate the concentration of the cccDNA in the peripheral blood leukocytes, and the relative effect of the treatment initiation, blood samples from 31 untreated and 67 treated patients were analyzed. The long-term treated patients with optimal viral suppression had no detectable HBV DNA in peripheral leukocytes, but the patients in the treatment group were selected among highly viremic patients having initiated antiviral treatment as naive (entecavir or tenofovir), within a year from sampling, thereby still displaying residual viremia. Unfortunately it was impossible to retrospectively determine the cccDNA content in the peripheral leukocytes in the basal samples from the treated patients, since they had been conserved as only plasma. The PBMCs and granulocytes were separately analyzed for the cccDNA content. The results are shown in Figure 4C. In the untreated patients, the cccDNA concentration in the PBMCs was significantly higher than in the granulocytes (P < 0.0001), which can generally only be detected in patients with viremia > 100000 IU/mL. In the patients undergoing treatment, the cccDNA was significantly reduced. This reduction was more evident in the PBMCs than in the granulocytes (P < 0.0001), and the difference between the PBMCs and granulocytes lost significance (P = 0.4571). The RI in the PBMCs and in the granulocytes was in line with that of the liver (2.7 and 2.5 log10, respectively).
5. Discussion
In this study, we described the development of a novel real-time PCR assay set for the quantification of HBV DNA in liver biopsy specimens or peripheral blood leukocytes from patients with HBV infections. These assays provide a specific method for the detection and quantification of both the total and cccDNA. An additional advantage of the proposed approach is the quantification of the single copy gene (2 copies per cell) hTERT as a measure of the cellular contents in the sample, which has a clear advantage over other cellular genes, such as actins, globins, etc. whose many different pseudogenes can have similar sequences. This is certainly more precise than the measurements based on the sample weight or total DNA concentration in the extract. In addition, the hTERT amplification is conditioned by the same extraction biases as the HBV templates (sometimes problematic when working with tissue), avoiding the need for an additional internal control (another benefit of this strategy). The specific cccDNA amplification is obtained by amplifying a region which spans across sequences that are interrupted or absent in the relaxed reverse transcribed templates on both the sense and antisense strands, similar to some proposed cccDNA assays (18-21) and different from others, which rely on only one single strand interruption (19, 20).
The hydrolysis probe real-time fast amplification kinetics minimizes the formation of template self-priming artifacts, allowing a high specificity for the cccDNA, even in the absence of DNAse treatment. In addition to the linear response to the standard curve, the reliability and accuracy of the quantification was also evaluated by a comparison with a well-established commercial assay. The results obtained by the “in house” HBV tDNA quantification method described here, and the COBAS AmpliPrep/COBAS TaqMan HBV test on the same samples, demonstrated a linear correlation over a wide range of concentrations. A slight difference in the absolute number of copies obtained may depend on the conversion IU to the copies (which is arbitrarily set by the producer of the commercial test, in the case of ROCHE: 5.82 copies per IU). Despite the slightly lower sensitivity in the plasma (15.2 vs. 9 UI/mL), compared to the commercial assay, the sensitivity of the novel assay is adequate for the detection and quantitation of HBV DNA in bioptic samples, even from patients treated with antiviral compounds for years.
The use of the same plasmid construct as a unique standard for all 3 assays (by definition isomolar) warrants the precise relative quantification of the 3 respective templates, thus allowing the reliable calculation of important biological parameters, such as the concentration of the cccDNA per 106 cells, the concentration of the tDNA per 106 cells, and the intrahepatic yield of the tDNA molecules per cccDNA molecule, an important measure of replicative capacity or index (RI). The performance of these real-time PCR assays for the HBV cccDNA and tDNA was evaluated in the DNA extracts from different biological specimens. Despite the limit of this study being the collection of biological samples from a provisional and not well characterized set of patients, in the absence of a long-term follow-up, the results obtained can be considered proof of certain concepts. The testing of liver biopsies by the described assays allowed the precise quantification of intrahepatic molecular parameters. Among these, the RI (derived parameter) can be considered the best direct marker of pure antiviral efficacy, in contrast to viral load in the peripheral blood, which is inevitably influenced by the cellular immune response, which clears infected hepatocytes, and by the humoral immune response, which clears viral particles from the bloodstream. The RI was determined in a small group of treated, virally suppressed patients, and compared with that of the untreated patients. In the former group, the RI appeared deeply depressed, and directly dependent on the activity of the reverse transcriptase. In the same biopsies, the absolute cccDNA concentration was reduced to a lesser extent, confirming that the HBsAg persistence in the face of antiviral therapy underlies the persistence of an unexpectedly vast pool of infected hepatocytes. Interestingly, this parameter may also be useful as a measure of effective intracellular antiviral activity in those patients who fail antiviral treatment in the absence of viral resistance mutations, a condition sometimes observed in patients treated with tenofovir.
Finally, the preliminary results obtained by using the assay on the peripheral blood leukocytes suggest that the cccDNA in the PBMCs might be an interesting marker of antiviral efficacy, as these cells have a shorter half-life than hepatocytes, and the cccDNA concentration responds promptly to antiviral treatment. In contrast, the cccDNA in the granulocytes, similar to the intrahepatic cccDNA, seems to be more slowly affected by treatment. Therefore, despite the cccDNA in the granulocytes being measurable only in highly viremic patients (> 105 IU/mL), further attention should be granted to this novel biomarker, as it may provide useful information on intrahepatic colonization, without the need for invasive procedures. Peripheral granulocytes may also be a source for the sequencing analysis of the intrahepatic virus (25).
In conclusion, a simple multiple template assay was developed for the quantification of the total and ccc HBV DNA in clinical samples, normalized to cellular content. This assay appears more versatile than previously described assays for cccDNA testing. Its straightforward design simplifies testing procedures, making it apt for extensive use in diagnostic labs. The use of this, as well as the other versatile tests (20) for the cccDNA quantification in the diagnostic setting, will provide more insight into the still obscure events in the conditions of the equilibrium of viral colonization/clearance in the liver of CHB patients in different stages of the infection, and a better interpretation of the changes induced by therapeutic interventions. Its use in longitudinal studies on a significant number of patients, with adequate follow-up, may allow a better understanding of the interplay between intrahepatic replication and the immune response, which influences peripheral viral output (HBsAg and HBV DNA) in the plasma and peripheral leukocytes. In view of functional cure strategies, novel virological biomarkers are needed, especially those that provide a more sophisticated interpretation of the simple old serological ones, such as quantitative HBsAg, HBeag, and anti- HBe.