1. Background
Hepatitis B virus (HBV) infection can cause infectious liver diseases, and about 400 million people throughout the world are chronically infected with this virus (1, 2), which has a high risk of progression to the development of liver failure, liver cirrhosis, and hepatocellular carcinoma (1). According to the similarity in the sequence of HBV, this virus is classified into eight genotypes and named using capital alphabet letters (A to H) (3). Recently, two other genotypes (I and J) were proposed for HBV (4, 5). The most common isolated HBV genotype in Iran is D (6).
The replication of HBV involves a unique process, which is the production of covalently closed circular DNA (cccDNA) from the HBV genome through the repair of relaxed circular DNA (rcDNA) in the nuclei of hepatocytes. The cccDNA acts as the template for viral RNA transcription that serves as a viral pregenomic messenger RNA (mRNA), or as messenger RNA coding for the envelope (S), polymerase, core, and X proteins (7).
Most of the antiviral agents have a profound effect on rcDNA whereas low or no effect on cccDNA (the episomal form of HBV) (8, 9). Relapse of HBV replication after discontinuation of antiviral therapy is not uncommon and may be the result of the persistence of viral cccDNA. It has been shown that monitoring of cccDNA of HBV in liver biopsy specimens may be a valuable marker for virus eradication (10).
Although detection and quantification of cccDNA in liver biopsy samples is the gold standard (11), it should be noted that liver biopsy is not always possible. Therefore, the detection of cccDNA should be performed on other samples, when a liver biopsy specimen is not available. Recent studies have indicated that the HBV cccDNA level is a marker of HBV replication in the hepatocytes of HBV-infected patients (12).
Hepatitis B surface antigen (HBsAg) level is an important marker of infection with HBV and it can be used to diagnose, manage, and monitor patients. Also, the level of HBsAg in the plasma of HBV-infected patients indirectly reflects the number of infected hepatocytes (13). It has been shown that the plasma HBsAg level was related to HBV DNA replication (7, 11). The hepatitis B surface antigen level has been proposed as a marker of infected liver or the amount of HBV cccDNA, which persists in hepatocytes (14).
2. Objectives
The present study aimed to detect HBV cccDNA in plasma sample of patients with HBeAg-negative chronic active hepatitis B and investigate the association between viral load and HBsAg level with the presence of cccDNA in plasma samples of the Iranian treatment-naive patients with chronic hepatitis B infection.
3. Patients and Methods
3.1. Study Population
From April 2012 to May 2015, 106 patients with chronic hepatitis B infection referred to Tehran Hepatitis Center and the hospitals that are related to Iran University of Medical Sciences were enrolled in this cross-sectional study. The participants were eligible if they had a positive HBsAg and a negative HBeAg for at least 6 months and were naive to antiviral treatment. The patients were excluded if they had coinfection with hepatitis C virus (HCV) or human immunodeficiency virus (HIV). This study was approved by the local ethics committee of the Gastrointestinal and Liver Disease Research Center (GILDRC) of Iran University of Medical Sciences, and all the patients provided written informed consent. A peripheral blood sample was collected from each patient in an EDTA-containing sterile tube and after separation of the plasma by centrifugation (5 minutes at 3000 rpm), the plasma samples were stored at -80°C for further experiments.
3.2. Hepatitis B Surface Antigen Quantification
The level of HBsAg was measured by the Roche HBsAg II assay on the Cobas e411 system (Roche Diagnostics GmbH, Mannheim, Germany), according to the manufacturer’s recommendations. This assay is based on chemiluminescent immunoassay (CLIA), which uses microparticles coated with monoclonal anti-HBs for the quantitation of HBsAg in plasma samples. This commercial CLIA has narrow dynamic range of quantification (0.05 - 130.00 IU/mL); therefore, samples that contain high levels of HBsAg must be retested after being diluted. If a result is found below the lower range, the specimen has to be run undiluted.
3.3. Hepatitis B Virus DNA Quantitation (Viral Load)
Hepatitis B virus DNA quantitation in the patients' plasma samples (500 μL) was performed using COBAS TaqMan 48 (Roche Diagnostics, Hacienda Drive Pleasanton, CA, USA) kit and high pure extraction was used according to the manufacturer’s recommendation. This assay is a real-time polymerase chain reaction (RT-PCR) method based on dual-labeled hybridization probe targeting the HBV precore and core regions. The detection limit of the COBAS TaqMan 48 is 6 > to 1 × 108 IU/mL.
3.4. Detection of Hepatitis B Virus Covalently Closed Circular DNA Using Real-Time Polymerase Chain Reaction
The viral DNA was extracted from 200 μL of plasma samples using QIAamp® DNA Mini Kit (Qiagen GmBH, Hilden, Germany), according to the kit instruction. To increase the specificity of cccDNA detection, plasmid-safe DNase (Epicentre, Madison, WI, USA) was used to eliminate rcDNA, single-stranded DNA (ssDNA), and replicative double-stranded DNA (dsDNA) prior to RT-PCR, based on the kit instructions.
The RT-PCR was performed for the detection of HBV cccDNA in plasma sample using the RT-PCR instrument, Rotor-Gene Q (QIAGEN, Germany), as described previously (15). Briefly, a pair of primers (the sense primer: 5′-ACTCTTGGACTCBCAGCAATG-3′; 1702 - 1722, and the antisense primer: 5′-CTTTATACGGGTCAATGTCCA-3′; 1962 - 1942) that can specifically amplify a DNA region from HBV cccDNA (not viral genomic DNA) were used (7, 15). For amplification of HBV cccDNA, 5 pmol of the TaqMan probe (5′-FAM-CTTTTTCACCTCTGCCTAATCATCTCWTGTTCA-TAMRA-3′; 1860 - 1892) and 10 pmol of each primer were used. The RT-PCR was performed using a 25-μL mixture containing 5 μL of the DNA template and 12.5 μL Maxima Probe qPCR Master Mix (Fermentas GmbH, St. Leon-Rot).
The cycling program of RT-PCR consisted of an initial denaturing step at 95°C for 10 minutes, followed by 45 amplification cycles at 95°C for 15 seconds and at 59°C for 1 minute. Extracted DNA from liver biopsy specimens of three patients with HBV infection, who gave consent and underwent a liver biopsy for diagnostic purpose, were used as positive controls for detection of HBV cccDNA. Liver biopsy samples were divided into 2 parts: one used for histological diagnosis, and the other was submerged into RNAlater (Ambion Inc., Austin, TX) and stored at -20°C.
3.5. Statistical Analysis
All statistical analyses were performed using SPSS software version 16.0 (SPSS 16.0 for Windows; SPSS Inc., Chicago, Illinois, USA). To find the normality of the data, the Kolmogorov-Smirnov test was used. Analysis of continuous variables was carried out using independent samples t-test or Mann-Whitney U test. The chi-square test or Fisher exact test was performed to assess associations between categorical variables. P Value of ≤ 0.05 was considered to be statistically significant.
4. Results
A total of 106 patients infected with chronic HBV were recruited in this cross-sectional study. The mean (SD) age of the study patients was 41.1 ± 12.4 years (age range, 20 - 62 years). From a total of 106 participants, 67 cases (63.2%) were males. The demographic characteristics and laboratory parameters of the Iranian HBeAg-negative patients with chronic hepatitis B infection are listed in Table 1, and the relationship between these parameters and the detection of cccDNA are shown in Table 2. The results of the Kolmogorov-Smirnov test showed that the distribution of HBV DNA levels and HBsAg titers was irregular and normal for age, aspartate aminotransferase (ALT), alanine aminotransferase (AST), and alkaline phosphatase (ALP).
| Variables | Patients | P Value | ||
|---|---|---|---|---|
| Male | Female | Total | ||
| Number of Subjects b | 67 (63.2) | 39 (36.8) | 106 (100) | |
| Age, y c | 41.42 ± 11.54 (21 - 64) | 44.28 ± 13.64 (20 - 63) | 41.1 ± 12.4 (20 - 62) | 0.274 (student t-test) |
| HBV Viral Load, IU/mL d | 21015.0 (652 - 7211488560) | 27435.0 (2065 - 498800) | 23055.0 (652 - 7211488560) | 0.331 (Mann-Whitney test) |
| HBsAg Titer, IU/mL c | 6622 ± 9203.17 (78-52300) | 5021.36 ± 7680.69 (7 - 28600) | 6033 ± 8671.25 (7 - 52300) | 0.362 (student t-test) |
| Baseline ALT Level, IU/L c | 32.02 ± 16.69 (10 - 80) | 35.11 ± 35.98 (10 - 167) | 33.17 ± 25.45 (10 - 167) | 0.672 (student t-test) |
| Baseline AST Level, IU/L c | 32.15 ± 16.61 (9 - 58) | 26.21 ± 13.47 (12 - 85) | 29.93 ± 15.68 (9 - 85) | 0.113 (student t-test) |
| Baseline ALP Level, IU/L c | 218.06 ± 76.8 (100 - 472) | 190.75 ± 39.47 (116 - 270) | 207.87 ± 65.90 (100 - 472) | 0.045 e (student t-test) |
| cccDNA b | 0.039 e (Fisher exact test) | |||
| Positive | 16 (23.9) | 3 (7.7) | 19 (17.9) | |
| Negative | 51 (76.1) | 36 (92.3) | 87 (82.1) | |
aAbbreviations: ALP: alkaline phosphatase; ALT: alanine aminotransferase; AST: aspartate aminotransferase; cccDNA: covalently closed circular DNA; HBV: hepatitis B virus; HBsAg: hepatitis B surface antigen.
bValues are presented as No. (%).
cContinuous parameters are presented as Mean ± Standard Deviation.
dMedian of HBV viral load: Because did not accept normal distribution in HBV viral load.
eStatistically significant.
| Variables | cccDNA | P Value | |
|---|---|---|---|
| Positive | Negative | ||
| Number of subjects b | 19 (17.9) | 87 (82.1) | |
| Gender-Female/Male | 3/16 | 36/51 | 0.039 (Fisher Exact) c |
| Age d | 44.16 ± 14.13 | 42.10 ± 12.0 | 0.514 (student t-test) |
| HBV Viral Load, IU/mL e | 899810.0 (652 - 7211488560) | 16866.0 (1612 - 1957360) | < 0.0001 (Mann - Whitney test) c |
| HBV Viral Load, IU/mL b | <0.0001 (Fisher Exact) c | ||
| < 60,000 | 4 (21.1) | 70 (80.5) | |
| ≥ 60,000 | 15 (78.9) | 17 (19.5) | |
| HBsAg Titer, IU/mL d | 10479.26 ± 13556.97 (169 - 52300) | 5062 ± 6937.36929.21 (7 - 28690) | 0.013 (student t-test) c |
| HBsAg Titer, IU/mL b | 0.041 (Fisher Exact) c | ||
| < 3000 | 5 (26.3) | 48 (55.2) | |
| ≥ 3000 | 14 (73.7) | 39 (44.8) | |
| Baseline ALT Level, IU/L d | 43.33 ± 10.45 (25 - 65) | 30.63 ± 27.46 (10 - 167) | 0.084 (student t-test) |
| Baseline AST Level, IU/L d | 41.80 ± 18.46 (19 - 85) | 26.97 ± 13.52 (9 - 73) | 0.001 (student t-test) c |
| Baseline ALP Level, IU/L d | 224.73 ± 73.93 (120 - 401.0) | 203.65 ± 63.72 (100 - 472) | 0.271 (student t-test) |
acccDNA: Covalently Closed Circular DNA; HBsAg: Hepatitis B Surface Antigen; ALT: Alanine aminotransferase; AST: Aspartate aminotransferase; ALP: Alkaline phosphatase.
bValues are presented as No. (%).
cStatistically Significant.
dContinuous parameters are presented as Mean ± Standard Deviation.
eMedian of HBV viral load: Because did not accept normal distribution in HBV viral load.
There was no statistically significant difference between age, laboratory parameters (ALT, AST, HBV viral load levels, and HBsAg titer), and the sex of the patients; however, there was a statistically significant difference between some laboratory parameters (ALP level [P = 0.045 with Student t-test]), and the presence of cccDNA in plasma sample (P = 0.039 with Fisher exact test) and the gender of the precipitants (Table 1).
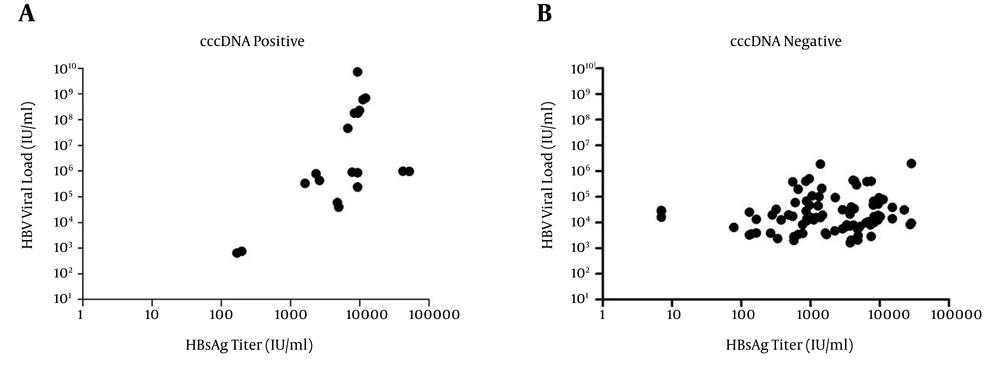
In the present study, there were 19 (17.9%) individuals with positive result for detection of cccDNA in plasma samples. A significant difference was seen in the HBV viral load levels between individuals with and without cccDNA in their plasma samples (P < 0.0001) and the presence of cccDNA in the plasma sample and high baseline HBV viral load (≥ 60,000 IU/mL) (P < 0.0001 with the Fisher exact test).
There was a meaningful correlation between the HBsAg titer and the presence of cccDNA in the patients plasma samples (P < 0.0043). Also, a statistically significant difference was observed between the presence of cccDNA in patients’ plasma specimens and high baseline HBsAg titer (≥ 3,000 IU/mL) (P = 0.041 with the Fisher exact test).
In the current study, there was a statistically significant difference between AST level and the presence of cccDNA in patients’ plasma specimens (P = 0.001). The HBV DNA levels and HBsAg titers between individuals with and without cccDNA in their plasma samples were shown in Figure 1.
5. Discussion
Hepatitis B virus infection manifests with a wide range of clinical symptoms; therefore, there is a need for sensitive and reliable markers to improve the management of this infection. Covalently closed circular DNA of HBV is responsible for viral persistence in the liver (16). There is little information about HBV cccDNA and its function in vivo (15). The HBV cccDNA level is a useful indicator for assessing the status of HBV replication in the liver, but its clinical use is extremely restricted because of invasive procedures during sampling (17). There are some reports indicating that the reason of chronic HBV infection is cccDNA (18). On the other hand, it has been demonstrated that cccDNA is the major cause for HBV reactivation after cessation of anti-HBV treatment (15). In this study, a significant association was seen between the HBV DNA level and the presence of cccDNA in the patients’ plasma samples (P < 0.000), and also between the HBsAg titer and the presence of cccDNA (P = 0.0043) in the patients’ plasma samples.
One important stage of the HBV life cycle is the production of HBV cccDNA, which serves as a template for HBV replication and plays a crucial role in the persistence of this viral infection (19, 20). It is suggested that the quantitation of intrahepatic HBV cccDNA is valuable and reliable for evaluating the efficiency of anti-HBV therapy (11, 21), and HBV recurrence after liver transplantation (22). It should be noted that this viral marker has not been applied in clinical practice widely (17).
The main disadvantage of detection of the HBV cccDNA before treatment response is the requirement for liver biopsies (16). However, it has been reported that the HBV cccDNA can also be detected in the plasma of HBV-infected patients (12, 15).
In the current study, a significant relationship was observed between the HBV viral load and the presence of cccDNA in the patients’ plasma. This finding is consistent with that reported by a study from Hong Kong (16) in HBeAg-negative patients.
It has been reported that serum cccDNA of HBV was detectable in 85.2% HBeAg positive and 48.1% negative chronic hepatitis B participants (12), whereas the HBV cccDNA was detected in plasma specimens in 19 (17.9%) out of the total 106 patients in the present study. Therefore, it seems that more research should be done on this area.
There was no statistically significant difference between the sex of the study population and age and laboratory parameters (ALT, AST, HBV viral load levels, and HBsAg titer) in the plasma samples; however, there was a statistically significant difference between the gender of the patients and some laboratory parameters (ALP level, and the presence of cccDNA). Shao et al. (23) showed that there was no relationship between the titer of HBV DNA and AST (P = 0.054), while plasma HBV DNA titer was correlated with ALT (P = 0.042) in HBeAg negative patients.
Chen et al. (15) showed a positive correlation between the sera cccDNA level and ALT level in patients with HBV reactivation, but not in individuals’ sera without HBV reactivation. Their results indicate that the occurrence of cccDNA in the sera is an early signal of liver damage (15). This finding is interesting and additional studies are required in this field.
Hepatitis B surface antigen is a marker of HBV infection, and serological tests for its detection have guided its diagnosis (24). The present study showed a positive correlation between plasma quantitative HBsAg and the presence of cccDNA in HBeAg-negative chronic hepatitis B participants (P = 0.041). This result is consistent with another study that was conducted among HBeAg-positive chronic hepatitis B individuals (16), but is different from the findings reported by studies from Greece (25) and Hong Kong (26).
It was shown that plasma HBsAg is a reflection of the amount of cccDNA when the level of cccDNA is high. However, the quantitation of HBV cccDNA is decreased by the immune clearance, and the percentage of cccDNA to HBsAg production with integrated HBV DNA, in the genome of hepatocytes, may be lower than that in individuals with HBeAg-positive chronic hepatitis B patients (27). Mechanisms that regulate the production of HBsAg from the integrated HBV DNA are not entirely clear. Thus, it is obvious that the production of HBsAg has no relationship with the quantity of the HBV template (cccDNA) and the replicative activity of the HBV in HBeAg-negative chronic hepatitis B individuals. Therefore, it seems that the quantity of the plasma HBsAg in participants with HBeAg-negative is not a reliable indicator of the HBV replicative efficiency (16).
In the current study, the HBV DNA, and HBsAg level detected in one session and may be repeating these tests will be more useful; however, we did not repeat them due to finance limitation.
In conclusion, the findings of the present study confirmed the concept that the plasma HBV viral load level and the quantitation of HBsAg have association with the presence of HBV cccDNA in the sera specimen. Therefore, it seems that detection of cccDNA in the plasma specimens is reliable and informative marker.